NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.
This publication is provided for historical reference only and the information may be out of date.
Scope
NCBI is a member of the Genome Reference Consortium (GRC), an international collaboration that oversees updates and improvements to the human, mouse, and zebrafish reference genome assemblies. These reference assemblies include linear chromosome representations, unlocalized and unplaced scaffold sequences, and alternate loci scaffolds providing alternate sequence representations for genome regions too complex to be adequately represented by the linear chromosome path. The GRC produces two types of assembly updates: (1) major releases, in which chromosome coordinates are changed, and (2) minor releases, in which chromosome coordinates do not change and updates are provided as standalone patch scaffold sequences. All GRC assemblies are submitted to the International Nucleotide Sequence Database Collaboration (INSDC) databases and made publicly available. The GRC is not responsible for annotation of the reference assemblies. For information about the National Center for Biotechnology Information’s (NCBI) annotation of the GRC assemblies, please see the handbook chapter titled, “About Eukaryotic Genome Processing and Tools”.
History
In 2004, the Human Genome Project (HGP) published a finished version (Build35) of the human genome assembly (1). This was a major accomplishment that represented over a decade of effort by more than a dozen institutions and resulted in the highest quality vertebrate genome ever produced and a new tool for understanding human biology. Despite this achievement, a limited number of gaps, sequence and tiling path errors remained in the reference assembly. Thus, at the conclusion of the HGP and the release of their final assembly version (Build36 (UCSC name: hg18)), the GRC was conceived as a mechanism for continued stewardship and improvement of the human reference assembly. The GRC was subsequently tasked with updating the mouse reference genome upon conclusion of its major sequencing effort and assembly release (MGSCv37) (2), and in 2010 the GRC also assumed responsibility of the zebrafish reference genome after the release of the Zv9 assembly.
The GRC is comprised of four institutions. NCBI supplies the database and provides bioinformatics support for the consortium, and also develops public-facing GRC assembly resources. Sequencing and other wet lab work associated with updating the assembly is performed by The Genome Institute at Washington University, St. Louis and at the Wellcome Trust Sanger Institute. The latter, along with the European Bioinformatics Institute (EBI) provide additional bioinformatics support and tool development for the GRC.
Although the GRC’s primary role was initially envisioned to be one of gap-filling and sequence correction, advances in genomic and population biology made possible by the availability of the human reference genome soon defined new assembly management tasks for the consortium. Notably, many studies of the human genome revealed previously unrecognized degrees and forms of genetic variation (3-10). The original assembly model, comprised of linear chromosome sequences, proved insufficient in its ability to represent this variation. Thus, the GRC, in addition to correcting assembly errors, also makes updates to the assembly model used to represent these organisms’ genomes and works to provide additional representations of diversity in the reference assemblies (11). In 2009, it produced an updated human assembly (GRCh37 (UCSC name: hg19)) and, in 2012, released a revised mouse assembly (GRCm38 (UCSC name: mm10)), the first two assemblies to be represented by the new model. Today, the GRC remains dedicated to producing improved reference assemblies that serve as valuable substrates for a variety of analyses.
Data Model
Assembly Model
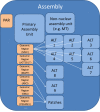
It is important to recognize that a genome assembly and a genome are not the same thing. A genome is the physical genetic entity that defines an organism. An assembly is not a physical object; it is the collection of all sequences used to represent the genome of an organism. The GRC utilizes a specific assembly model for the reference genomes under its auspices (Figure 1). However, this assembly model can be adopted for use with almost any eukaryotic genome. Within this model, sequences belong to different hierarchies and are assigned to various assembly units, depending upon their role in assembly.
Sequence Hierarchies
Because current sequencing technologies do not allow for chromosomes to be sequenced from end-to-end in a continuous fashion, they must be fragmented, sequenced, and reassembled for purposes of representation. The minimal collection of sequences needed to reconstruct a molecule of interest is referred to as its tiling path. The reference assembly model includes three tiers of accessioned sequences. Figure 2 uses human chromosome 6 (CM000668.1) to illustrate this hierarchy. At the bottom of this hierarchy are the tiling path components, which in the case of the GRC reference assemblies are primarily genomic clones or Whole Genome Shotgun (WGS) contigs. In the middle are scaffolds, which are sets of ordered and oriented components. At the top of this hierarchy lie the chromosome sequences. These are assembled from scaffolds that have been localized and oriented with respect to one another and that are separated from one another by gaps representing unresolved sequence. A genome assembly may also contain scaffold sequences whose chromosomal context is either poorly defined or not known. The former category describes unlocalized scaffolds. These are genomic sequences that have been assigned to a particular chromosome, but whose location within that chromosome cannot be unambiguously defined at this time. Scaffolds entirely without chromosomal context are known as unplaced scaffolds.
Primary Assembly Unit
The primary assembly unit is the collection of sequences that, all together, provide a haploid representation of an organism’s genome. Prior to the development of this assembly model, the human reference assembly only consisted of the sequences in the primary assembly unit. As a result, researchers sometimes mistakenly continue to refer to the collection of sequences in the primary assembly unit as the reference assembly. However, this is only one of several assembly units that together comprise GRC assemblies.
The primary assembly unit includes the chromosome sequences and the collection of unlocalized and unplaced scaffolds. These scaffold sequences make important contributions to the primary assembly unit. For example, in the GRCh37 primary assembly unit, an unlocalized scaffold associated with chromosome 1 provided the only representation for the HYDIN2 locus (GL000192.1). Although this locus is known to reside on chromosome 1, a complex repeat structure confounded the chromosome assembly and made the assignment of this scaffold to any one of three gaps equally likely. Consequently, the scaffold was designated unlocalized.
Alternate Loci Assembly Units
Alternate loci assembly units contain sequences that represent variants of sequence present in the primary assembly unit. As such, they permit an assembly to provide more than a haploid representation of a genome. While there are no size limits for sequences in alternate loci assembly units, these are generally scaffold sequences less than 5 Mb in length. In the human reference assembly, which does not represent an individual genome, alternate assembly units are not organized by haplotype. In contrast, alternate assembly units in the mouse reference assembly are organized by strain; they only include sequences from strains other than C57BL/6J, which is represented in the primary assembly unit. No alternate assemblies have yet been defined for the zebrafish reference assembly. For GRCh37, the GRC instantiated 7 alternate loci assembly units so that the reference assembly might better represent the diversity that exists in the major histocompatibility complex (MHC) region on human chromosome 6, one of the most variable regions of the human genome (Figure 3). There are therefore 8 sequence representations for the MHC in GRCh37: one on the chromosome sequence from the primary assembly unit (CM000668.1), and 7 from scaffolds belonging to 7 alternate loci assembly units (GL000250.1-GL000256.1).
Patches Assembly Unit
All patches belong to the patches assembly unit. Patches are scaffold sequences that represent updates made to the reference assembly since its last major release. Thus, the patches assembly unit is empty at the time of an assembly’s major release. The GRC releases patches on a quarterly basis; the patches assembly unit always contains the complete collection of patches associated with the reference assembly. Patches do not change the coordinates of any sequences in the primary assembly or alternate loci units. The assembly model includes the concept of patches because they provide a mechanism for providing users with timely access to assembly improvements without the need for frequent major assembly releases involving chromosome coordinates updates that many researchers find disruptive. The GRC does not integrate the patch scaffolds into the chromosomes; they exist only as scaffold sequences.
There are two types of patch scaffolds in this assembly unit. Fix patches correct errors in the primary and alternate loci assembly units, while novel patches add new sequence variants to the assembly. As illustrated in Figure 4, the fix patch GL339450.1 provides a single haplotype representation for the ABO locus, correcting the mixed, non-existent haplotype found in GRCh37 where the locus spanned two components with different haplotypes. In Figure 5, the novel patch GL383583.1 is shown to represent a deletion variant involving the APOBEC3A and APOBEC3B genes, which are involved in innate immunity and retroviral infections. The deletion variant, which is common in Asia but rare in Europe and Africa, creates a gene fusion, APOBEC3A_B (12). At the time of an assembly’s next major release, all fix patch scaffold sequences will be deprecated, as the changes they represent will be reflected in sequences in the primary assembly and alternate loci assembly units. In contrast, novel patch scaffold sequences will be retained, though they will be moved from the patches assembly unit to the appropriate alternate loci assembly unit.
Non-Nuclear Assembly Unit
Although the GRC is not responsible for the maintenance of the mitochondrial reference sequences of the human, mouse, or zebrafish genomes, the assembly model includes a unit for non-nuclear assemblies. The human mitochondrial reference sequence is maintained by the Mitomap group and is distributed by the GRC with the reference genome assembly for user convenience.
Alignments
Although scaffolds in the patches and alternate assembly units do not have chromosome coordinates, they may be placed in chromosome context by virtue of their alignment to primary assembly sequences. All patch scaffolds and scaffolds in the human alternate assembly units contain at least one anchor sequence as either the first and/or last component (Figures 4 and 5). These anchor sequences are components that are also found in the primary assembly unit and are included to ensure a good alignment of the alternate locus scaffold to the primary assembly. Because the alternate loci assembly units in the mouse assembly are strain specific, their scaffolds do not contain anchor sequences from the primary assembly unit. As a result, mouse alternate loci scaffolds may not always have an alignment to the primary assembly unit.
The GRC generates alignments of the alternate loci and patch scaffolds to the primary assembly unit and submits these alignments to the NCBI Assembly http://www.ncbi.nlm.nih.gov/assembly/database with every assembly release. As a result, these alignments are part of the assembly definition and are distributed on the GenBank FTP site with the assembly sequences. The alignments distinguish how scaffold sequences from the patches or alternate loci assembly units differ from the primary assembly unit sequence. Figures 4 and 5 also show the alignments between the annotated RefSeq copies of the aforementioned fix and novel patches, and the corresponding GRCh37 chromosome sequences.
Assembly Regions
The GRC defines discrete regions on sequences in the primary assembly unit where alternate loci and patch scaffolds are aligned. A region may contain more than one patch or alternate loci scaffold and the extent of a region is defined by the outermost edges of the corresponding alignments. The GRC also defines regions on the X and Y chromosomes corresponding to the extents of the pseudo-autosomal regions (PAR), as defined by their alignments to one another. The ideogram in Figure 3 shows the location of regions associated with the GRCh37 assembly.
Assembly Accessions
All GRC assembly sequences are submitted to GenBank and the assembly itself is submitted to the NCBI Assembly database. Every scaffold and chromosome in the assembly receives an accession.version, which is a unique identifier of the sequence. Likewise, the assembly units and full assembly also receive accession.versions. These identifiers enable users to track the collections of sequences within each assembly. The GRC strongly recommends that authors include the accession.versions of all assembly sequences referenced in their publications. Because sequence coordinates may change with each accession.version update, use of these identifiers provides an unambiguous definition of the coordinate-sequence relationship. Such usage eliminates any possible reader confusion with respect to the particular sequence on which coordinates may be reported for genes, regulatory regions or other assembly features.
Dataflow
Figure 6 provides a schematic of the GRC dataflow for assembly updates. GRC assemblies start with a set of text files known as TPFs (tiling path files). TPFs provide an ordered list of the components and gaps that make up a scaffold or chromosome. However, they specify neither the orientation of the components, nor the specific sub-regions of the components that will contribute to the final sequence. GRC curators download TPF files from an NCBI database and update them with changes to the tiling path by adding, removing, or reordering components as indicated by their analyses. All updates are made in accordance with a series of GRC-developed standard operating procedures for assembly curation and the GRC uses a centralized system to track the regions of the assembly under review. The TPF files are then reloaded to the database, where they are validated for format and content. A versioning system ensures that all TPF updates are recorded, and a check-in/check-out system for the files prevents simultaneous modification of a TPF by more than one curator.
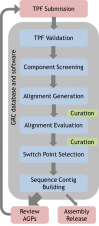
Figure 6.
Dataflow for GRC assembly updates.
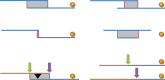
A modified version of the NCBI NGAligner software identifies and evaluates alignments between adjacent components with respect to criteria such as length and percent identity. Adjacent assembly components are generally expected to have dovetail overlaps (Figure 7), though other alignment types are sometimes observed. Pairs without alignments or those whose alignments do not meet established GRC evaluation criteria are prioritized for review. There are three possible outcomes of review: (1) the TPF may be further updated to solve the problem, (2) a new alignment meeting evaluation criteria may be curated and stored, or (3) the GRC may provide external evidence supporting the pairing of the sequences despite the low quality alignment (join certification). If a pair exhibits more than one alignment, a curator will designate the preferred alignment. The pairwise alignments and evaluation results are stored to the database. As a result, alignments need only be generated and evaluated for new sequence pairs on new or updated TPFs.
NCBI-developed software is also used to select switch points for each aligned pair (Figure 7). The switch points define the start and stop positions of the individual components in the scaffolds. By default, this occurs at the last base of the first component in the aligned pair. If an alignment does not exhibit 100% identity, which may occur when components represent different haplotypes or other forms of variation, the GRC may curate the switch points in order to include or exclude sequence unique to one of the components. Like the alignments, switch points are stored in the database and are only generated for new sequence pairs on new or updated TPFs. All switch points are validated to ensure they occur at aligned bases.
NCBI sequence contig building software known as tpf_builder uses the component order specified on the TPFs and the stored alignments and switch points to build sequence contigs and generate AGP (A Golden Path) files that describe the assembly scaffolds and chromosomes (Figure 6). During the inter-release period for an assembly, this software runs every time there is a sequence-changing TPF update. Any errors encountered in the process are reported to curators for their review, and the entire assembly curation process is repeated as necessary. At the time of a public assembly release, tpf_builder is triggered to produce a final set of AGP files. The alignments of the patch and alternate loci scaffold alignments to the primary assembly are also produced at this time, as are the genomic region definitions. These files are submitted to the NCBI GenColl database and subsequently loaded to GenBank, culminating in an assembly release.
There are two types of assembly releases. Minor releases are used by the GRC for updates to the patches assembly unit. In a minor release, the accession.version of the patches assembly unit and the full assembly will increment. However, the accession.version of the primary assembly unit and the alternate loci subunits will not change. As a result, there are no changes to the sequences or of any of the assembly chromosomes. In a major assembly release, all assembly unit accession.versions will increment. Major assembly releases are associated with coordinate changing chromosome updates. Users can distinguish whether a new GRC assembly represents a major or minor release by comparing the accession.version of the primary assembly unit in the latest assembly version to that of the previous assembly version: if the version is unchanged, it is a minor release; if it has incremented, it is a major release. Users can find accession.version information for all GRC assemblies in the NCBI Assembly resource.
Access
Users can download GRC assembly data from the GenBank FTP site. This data includes the sequences, alignments, assembly region definitions, and join certifications. The genome browsers at UCSC, Ensembl and NCBI, which obtain the assembly data from GenBank, provide displays for the GRC assemblies. The GRC generates a file that provides the genomic locations for all issues under review, which Ensembl and UCSC display as a track in their browsers. All three browsers have tracks showing the regions in the primary assembly for which there are patch and alternate loci scaffold sequences.
The GRC provides users with access to the inter-assembly TPF and AGP files on the GRC FTP site. While these files are not recommended for publication-level analyses, due to their instability and lack of corresponding accessioned sequences, they provide users with a preview of genome changes. At this FTP site, the GRC provides a file with the genomic locations of annotated clone assembly problems in the component sequences, which can also be loaded as a browser track.
The GRC strives to makes its efforts to update the human, mouse, and zebrafish reference assemblies as transparent as possible. It maintains a public website (Figure 8) where users can find assembly statistics for current and past assembly releases, plans for future updates, and a link to the GRC blog. At the GRC website, users will find pages describing the current status and genomic locations of individual issues under GRC review (Figure 9). Users can search the GRC website for specific issues by features such as genome location, gene name, accession, or clone name, and links are provided to view the corresponding regions in the major browsers. Additionally, the GRC website includes region-centric pages that provide links to the issue reports and sequence records for all patches, alternate loci, and issue reports associated with a specified region, along with a graphical view of the region (Figure 10). The website also provides forms for users to report assembly issues directly to the GRC, which are entered into the GRC tracking system, as well as to contact the GRC with general assembly questions.

Figure 8.
GRC website. A: GRC announcements. B: Link to and highlights from GRC blog. C: Link to and highlights of recently resolved assembly issues.

Figure 10.
Screenshot of NCBI Sviewer display from the GRC region-specific page for the human major histocompatibility complex (MHC) region on chromosome 6. This Sviewer instance includes several default tracks useful for evaluation of the chromosome or scaffolds (more...)
The GRC also provides users with access to the evaluated alignments, switch points, and join certificates for all sequence pairs on the assembly TPFs (Figure 11). Users can search for specific TPFs by component accession or clone name. The TPF Overview pages present an enhanced view of the TPF files that includes information such as the evaluation status, length, and percent identity for all component alignments. The OverlapView pages, accessed by clicking on the evaluation status markers in the TPF Overview pages, provide alignment and switch point details for each sequence pair in graphical and text formats. There is a link on each OverlapView page that can be used to view the alignment in Genome Workbench. The OverlapView pages provide information about the database history for the sequence pair, genomic clones whose ends map to either of the components, as well as the coordinates of RepeatMasked regions within the alignment. Links to pages showing join certificates submitted by GRC curators are found in the OverlapView pages for sequence pairs with sub-optimal alignments.
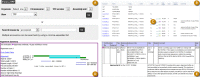
Figure 11.
A: Search interface for TPF pages. B: Detail from TPFOverview page for the mouse chromosome 16 TPF showing enhanced TPF file display table. Clicking on any join evaluation icon (C) will take a user to the OverlapView page for the specified pair. D: Detail (more...)
Related Tools
MapViewer and Sviewer
Users can view GRC assemblies and sequences in the NCBI MapViewer and Sviewer resources. These resources can be configured to show different tracks containing assembly data.
Clone DB
The NCBI Clone DB maintains records for the genomic clones that are components of the GRC assemblies, as well as for other, non-component clones. These records include sequence, distributor, and mapping information.
Genome Remapping Service
The NCBI genome remapping service can be used to remap features between different assembly versions.
Eukaryotic Genome Annotation Pipeline
All GRC assemblies are annotated as part of NCBI’s eukaryotic genome annotation pipeline.
References
- 1.
- Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45. [PubMed: 15496913]
- 2.
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS biology. 2009;7(5):e1000112. [PMC free article: PMC2680341] [PubMed: 19468303]
- 3.
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nature genetics. 2004;36(9):949–51. [PubMed: 15286789]
- 4.
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nature genetics. 2006;38(1):75–81. [PubMed: 16327808]
- 5.
- Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nature genetics. 2006;38(1):82–5. [PubMed: 16327809]
- 6.
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, et al. Fine-scale structural variation of the human genome. Nature genetics. 2005;37(7):727–32. [PubMed: 15895083]
- 7.
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome research. 2006;16(9):1182–90. [PMC free article: PMC1557762] [PubMed: 16902084]
- 8.
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–6. [PMC free article: PMC2674581] [PubMed: 17901297]
- 9.
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. [PMC free article: PMC2424287] [PubMed: 18451855]
- 10.
- Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, et al. Segmental duplications and copy-number variation in the human genome. American journal of human genetics. 2005;77(1):78–88. [PMC free article: PMC1226196] [PubMed: 15918152]
- 11.
- Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, et al. Modernizing reference genome assemblies. PLoS biology. 2011;9(7):e1001091. [PMC free article: PMC3130012] [PubMed: 21750661]
- 12.
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS genetics. 2007;3(4):e63. [PMC free article: PMC1853121] [PubMed: 17447845]
- Genome Reference Consortium - The NCBI HandbookGenome Reference Consortium - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...