Help
Welcome to NCBI Virus!
This page will help you to get started. It will guide you through the resource pages and explain available functionalities.
Please contact us if you have further questions.
How To
-
NCBI Virus is an integrative, value-added resource supporting retrieval, display and analysis of a curated collection of virus sequences and large sequence datasets. Find more about our data model.
What is NCBI Virus?
Main functionalities
-
Compare your sequence to those in the NCBI Virus database using NCBI BLAST algorithm. Learn more.
-
Search, view and download nucleotide and protein sequences using virus name or taxonomy group. Learn more.
-
Quickly access common data sets for all viruses, all human viruses, bacteriophages, or sequences released in the past month. Learn more.
-
Explore the massive, normalized datasets and identify data trends. Learn more.
Ways to access NCBI Virus data
Select one of the three options to access NCBI Virus data.
Option 1:
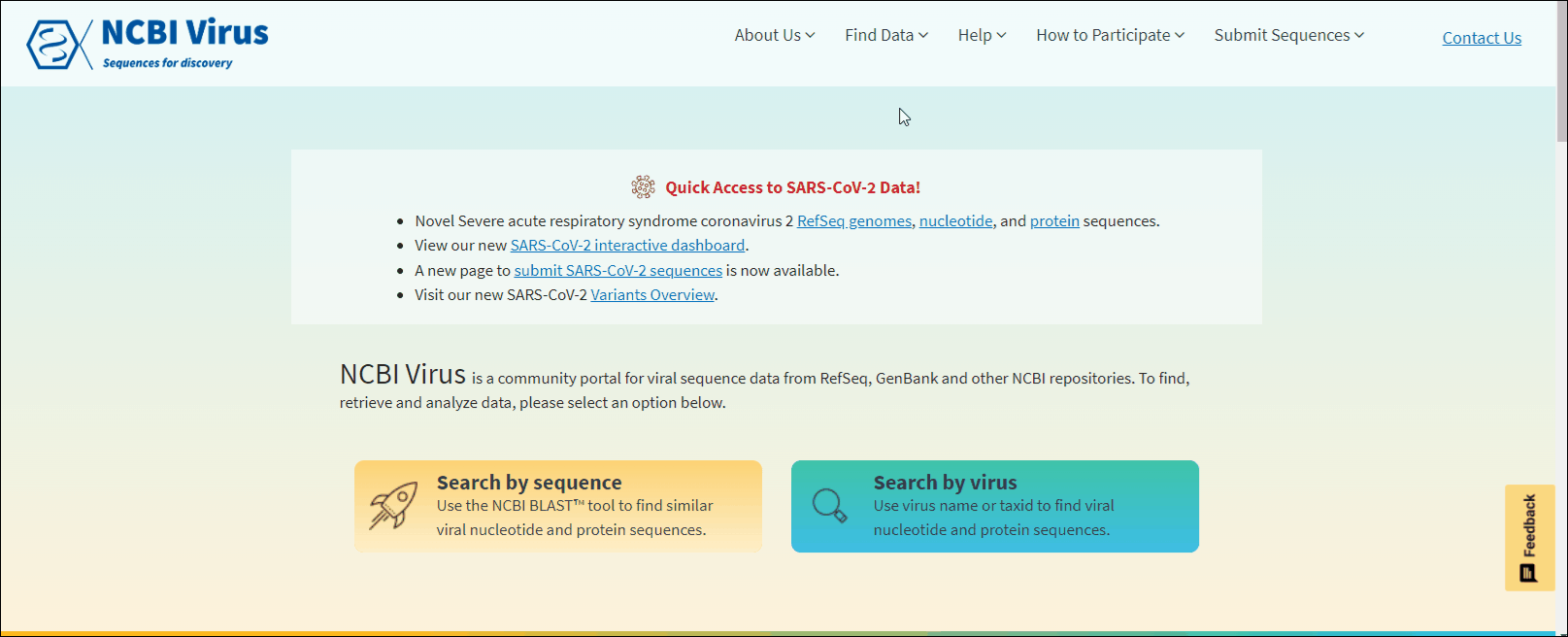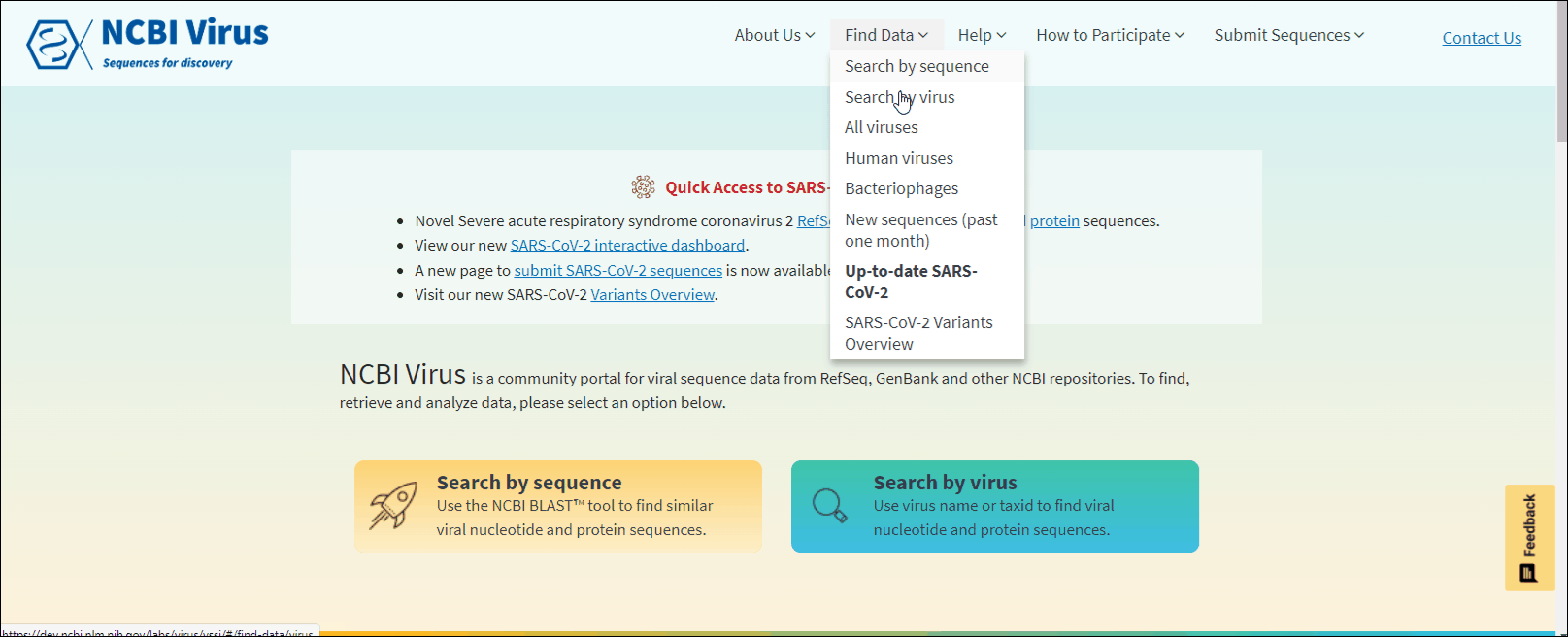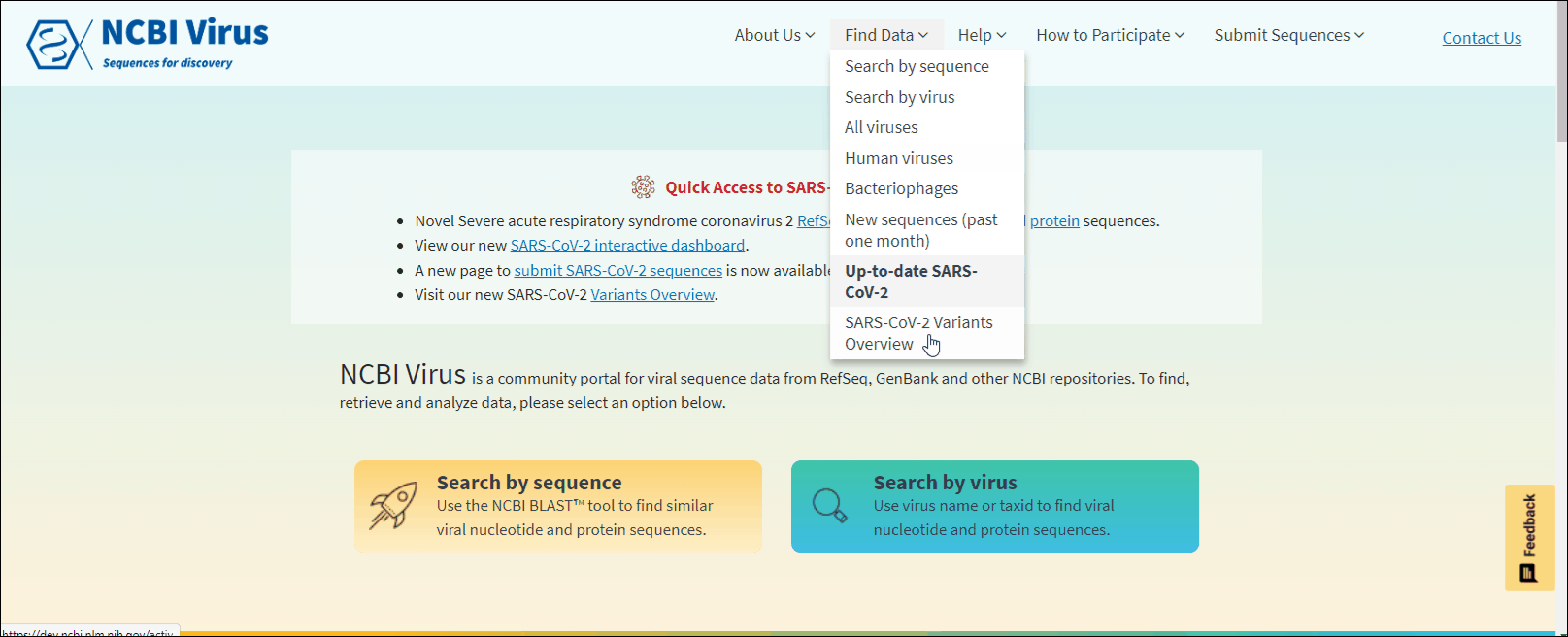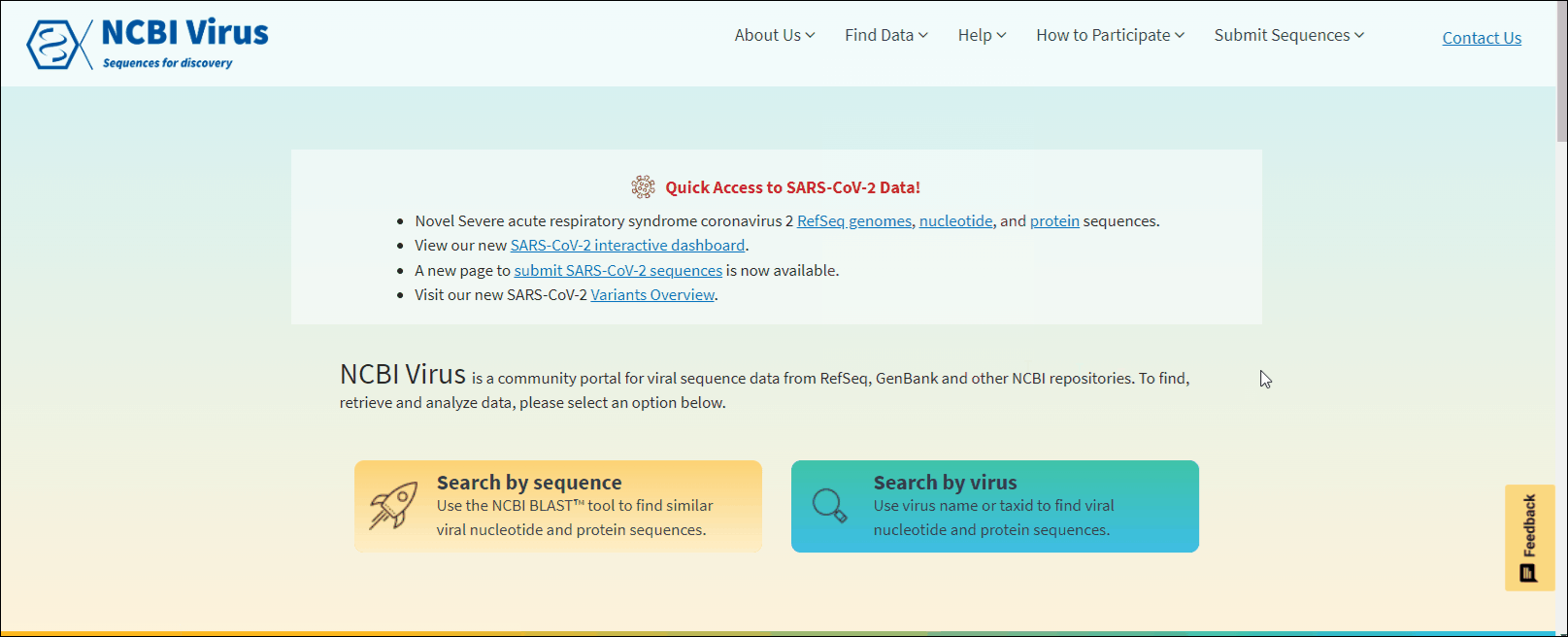
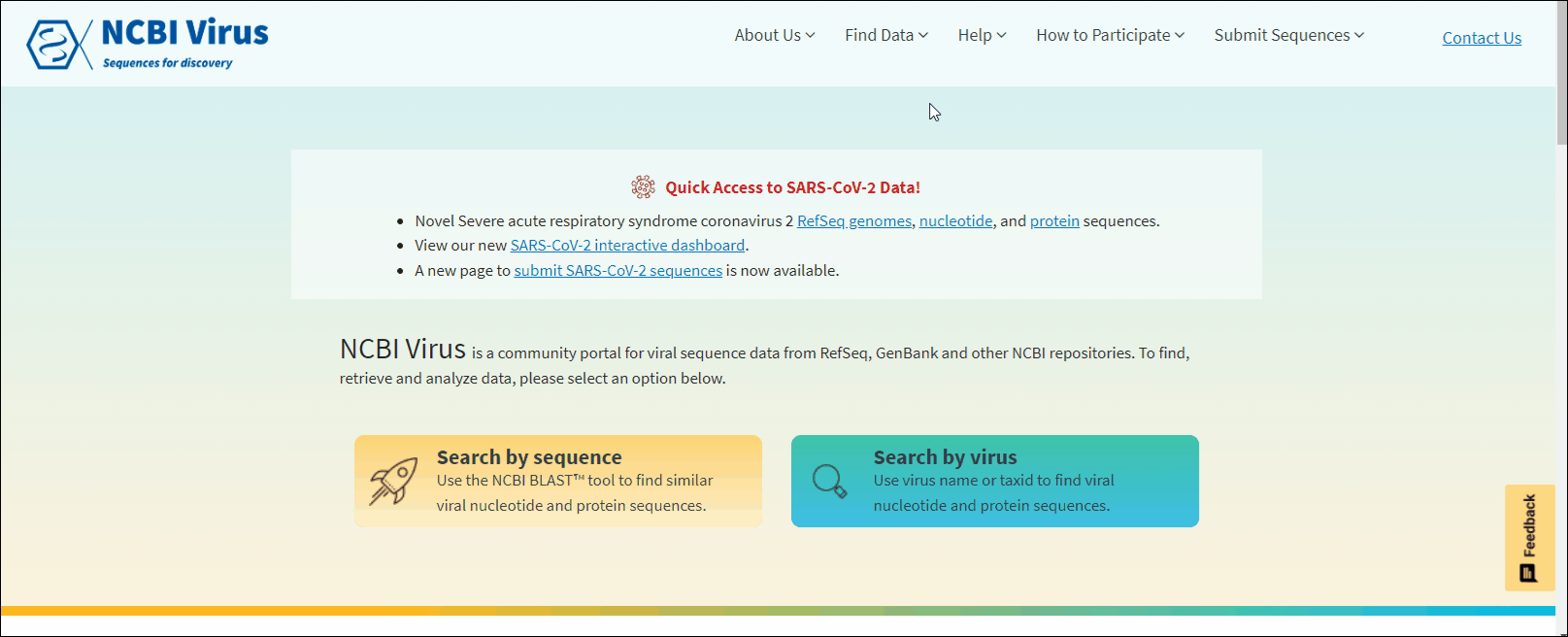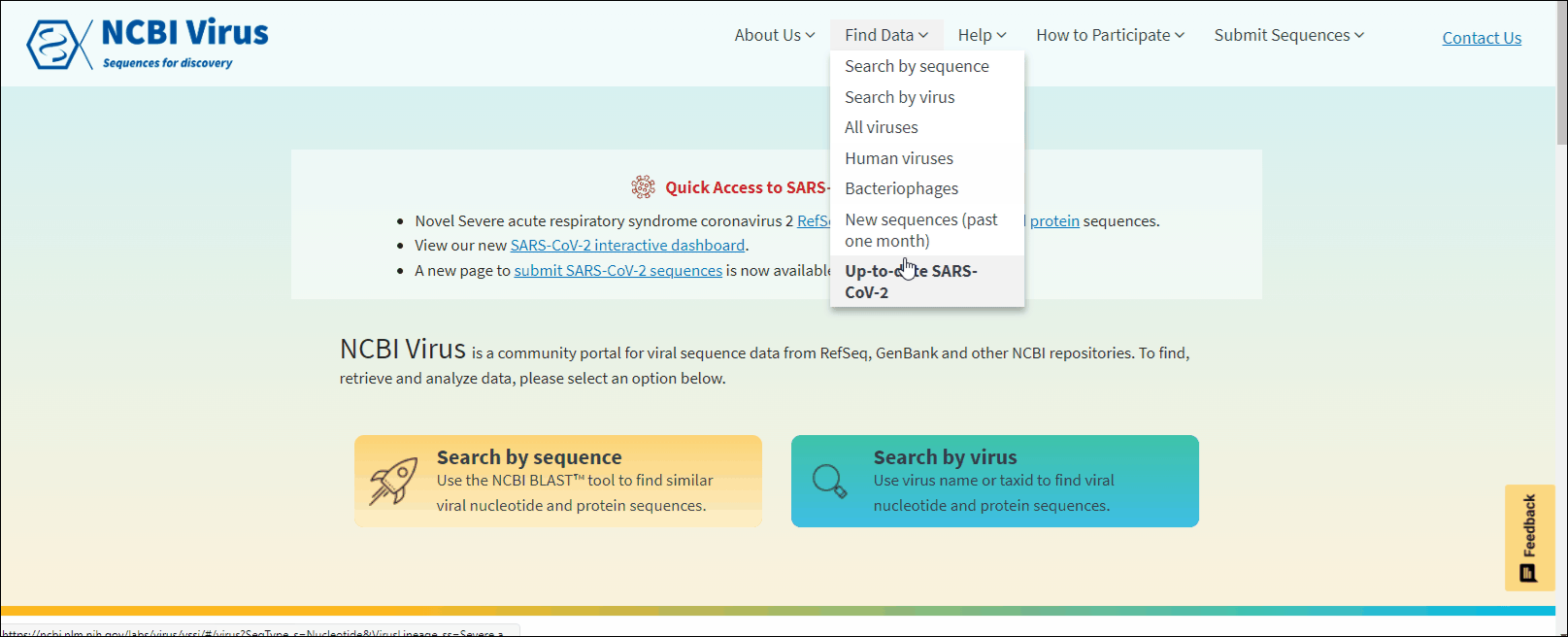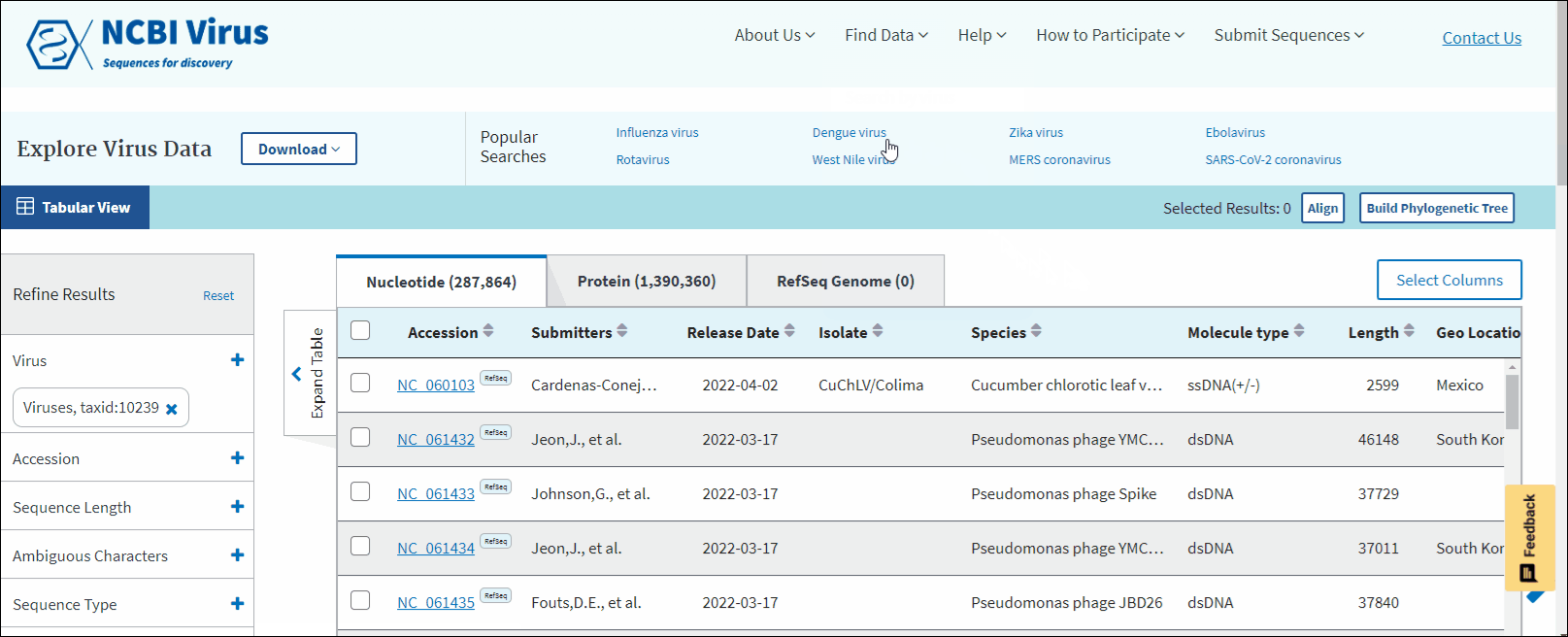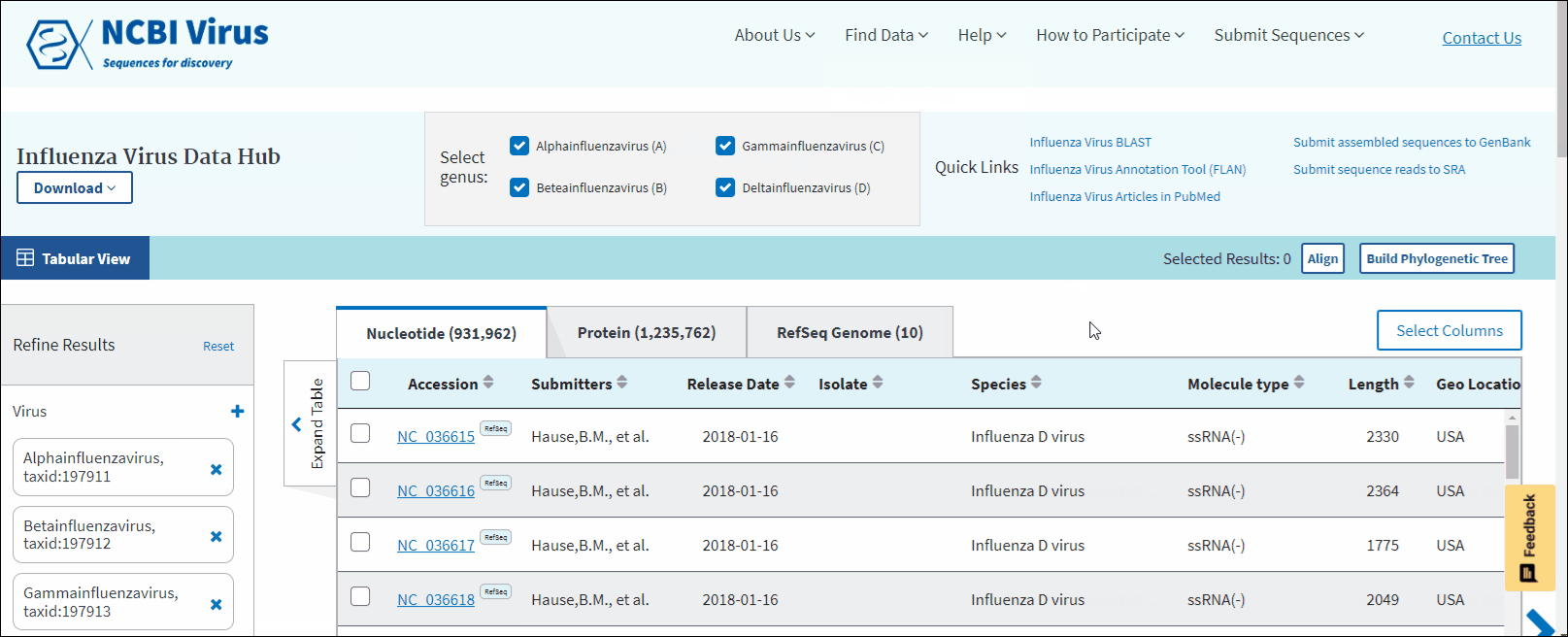
Through the navigation menu in Find data tab select one of the drop-down links:
-
Search by sequence to use virus-specific NCBI BLAST tool. Learn more.
-
Search by virus to perform virus sequence search based on virus name or taxonomy. Learn more.
-
All viruses, Human viruses, Bacteriophages, New sequences (past one month) and Available SARS-CoV-2 sequences to view preselected data sets. Learn more.
Option 2:
The same functionalities can be accessed through the buttons Search by sequence and Search by virus located on NCBI Virus home page.
The results can be viewed in the Results Table, and further refined by utilizing the sequence attributes (metadata) in the Refine Results panel located on the right side of the table. Additionally, you can download the results, conduct multiple sequence alignments, and generate phylogenetic trees using the selected results.
Learn more about BLAST search Results Table and virus name/taxonomy based search results.
Option 3:
Through NCBI Visual Data Dashboard via statistics buttons located in the top row of the dashboard. Learn more.
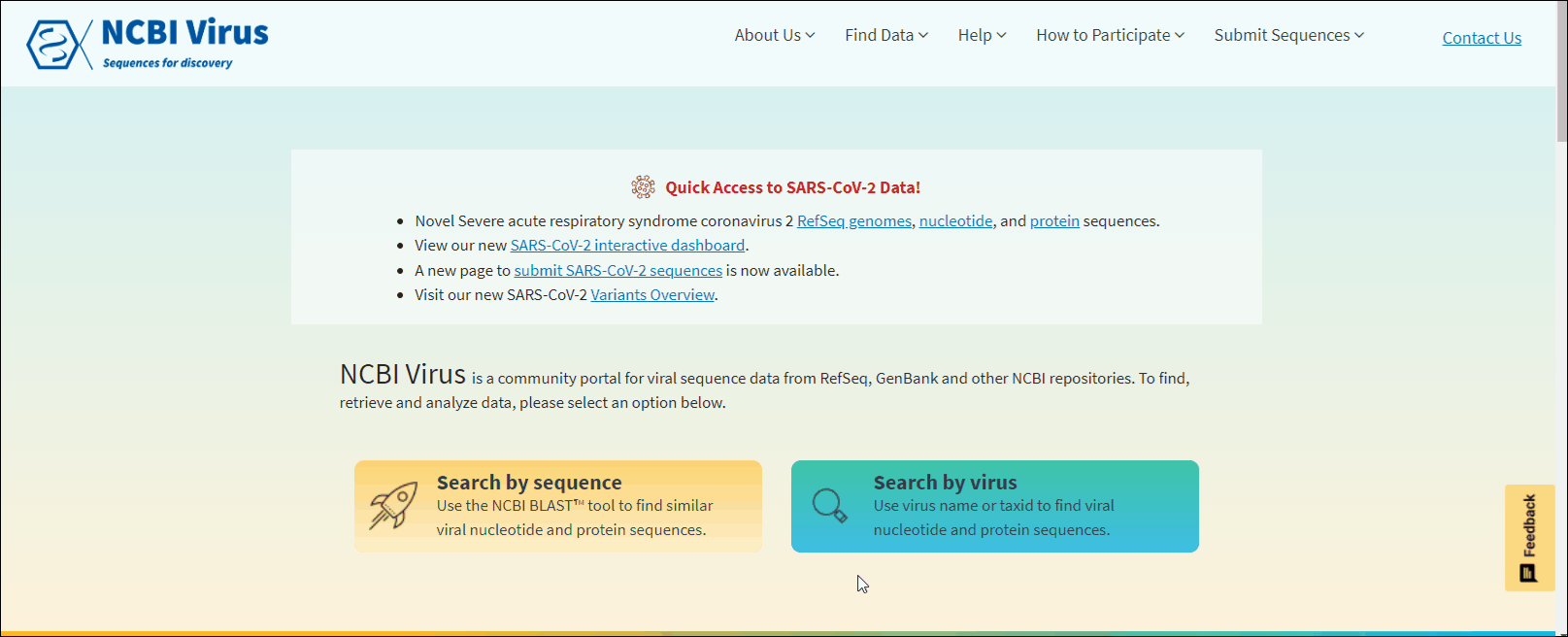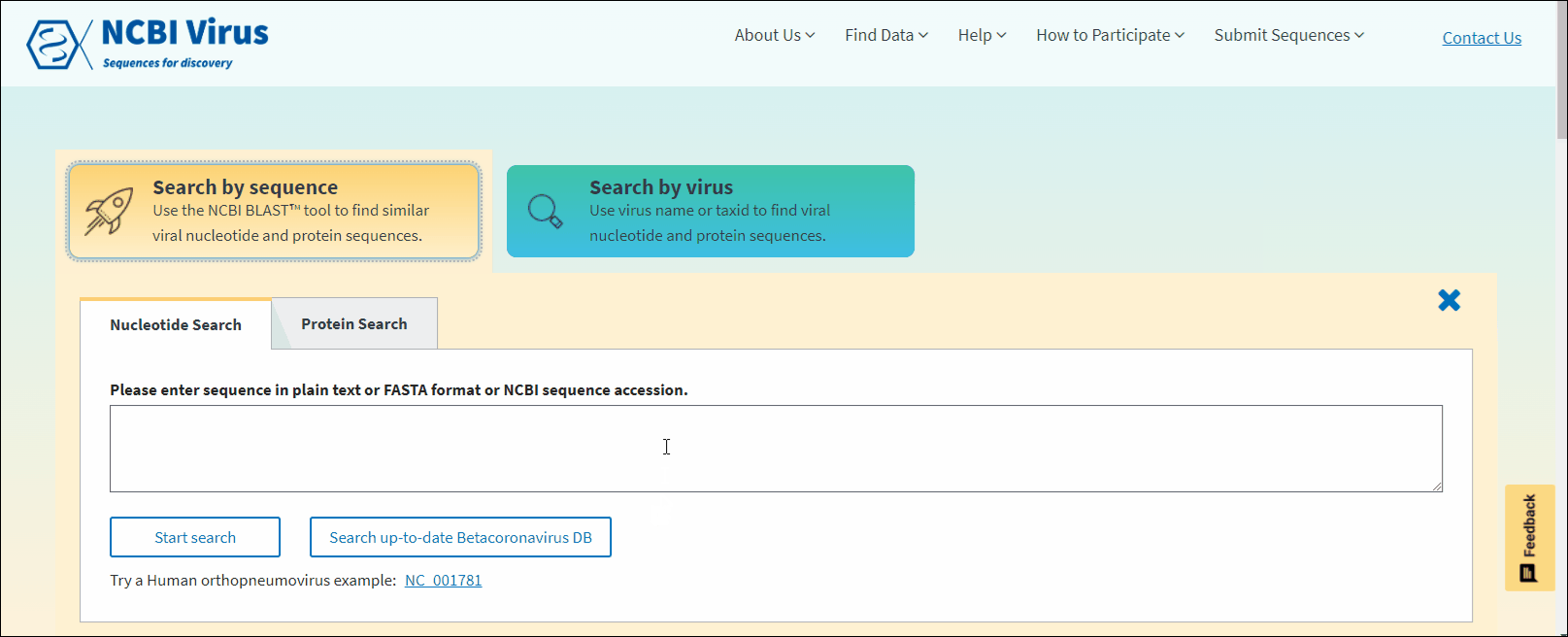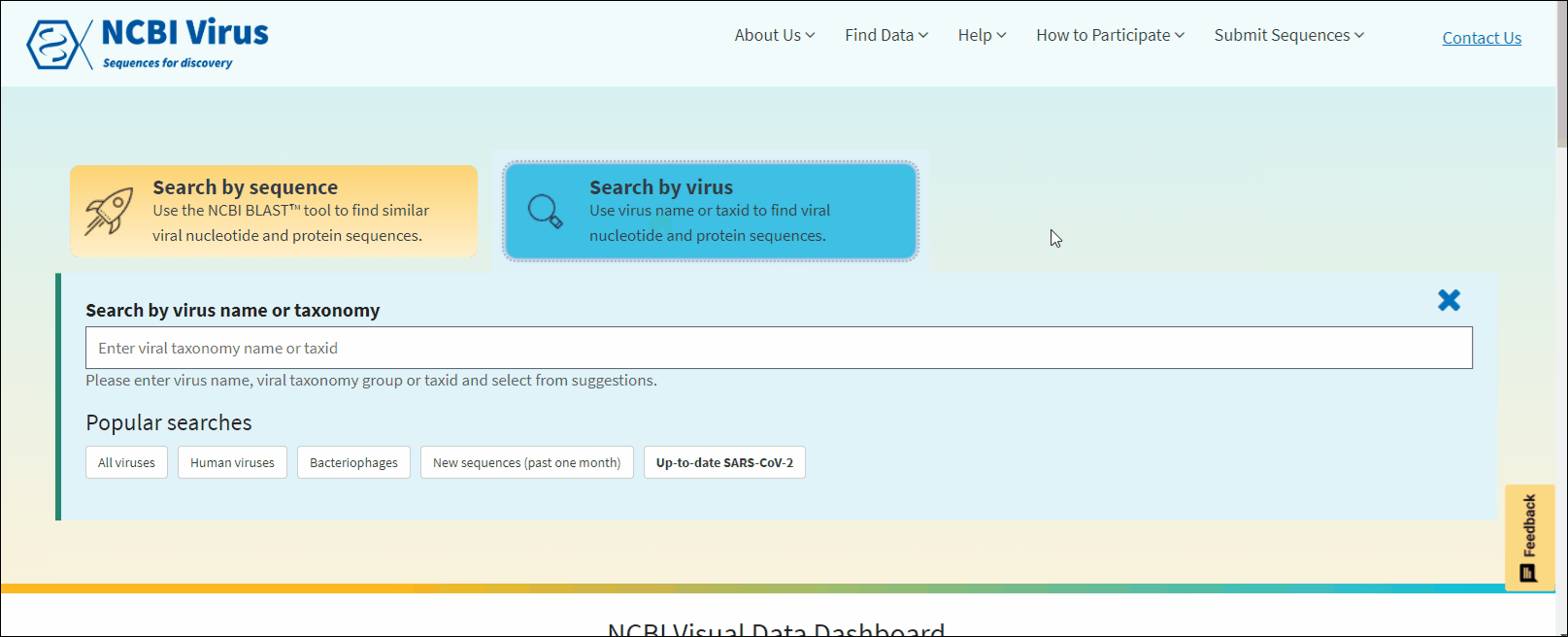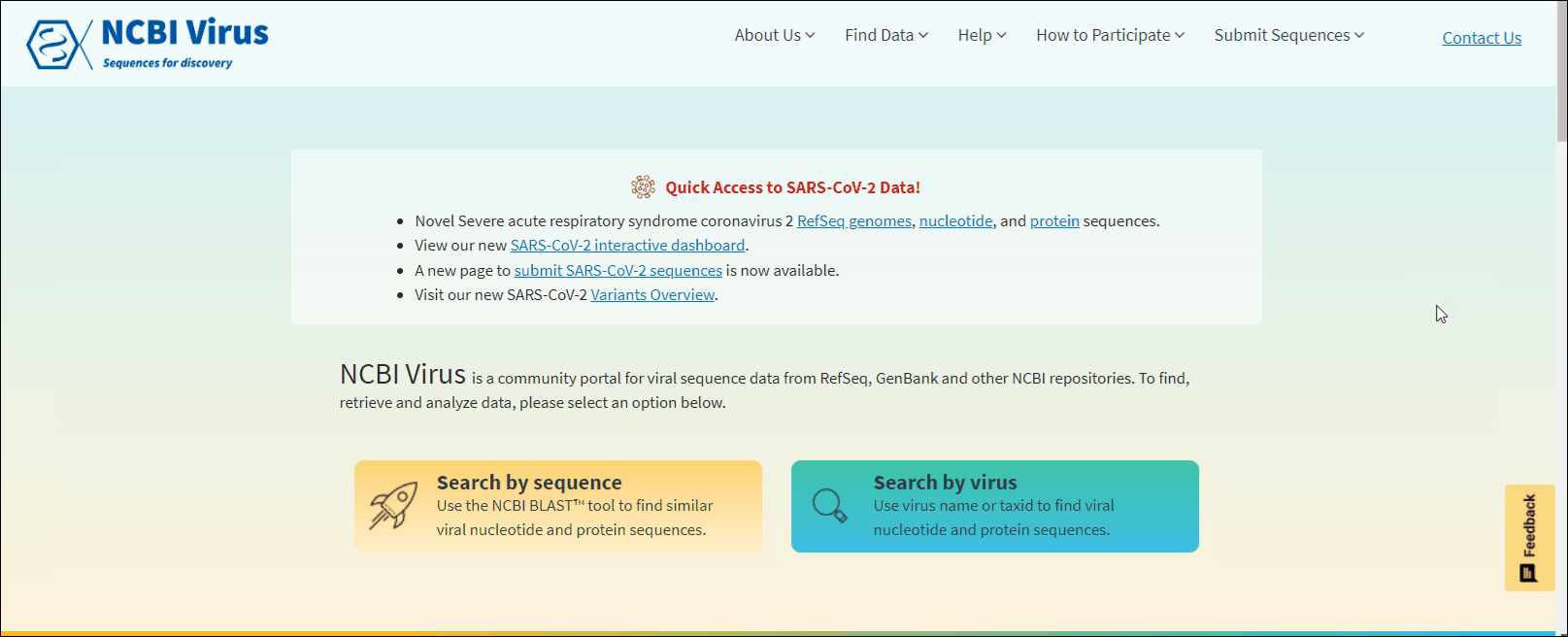
NCBI Virus BLAST™ tool
The NCBI Virus BLAST™ tool provides rapid insight into query sequences by presenting BLASTn and BLASTp results alongside normalized metadata, when available. These attributes include: isolation source, host, country, collection and release date, as well as taxonomy and genetic attributes such as completeness, and segment or protein names when applicable. The normalized metadata is generated via an internal, curator-guided data-processing pipeline that maps sequence-record attributes to standardized vocabularies to provide a user-friendly view of the data.
Compare your sequence to those in the NCBI Virus database using the BLAST algorithm
-
Press on the button Search by sequence (or select this option from the Find data navigation tab on the top of the page).
-
Select Nucleotide or Protein tab. Nucleotide tab allows to perform BLASTn search (search against all NCBI virus nucleotide sequences). Protein tab allows to perform BLASTp search (search against all NCBI virus protein sequences). Read more about BLAST™ searches at NCBI BLAST Guide.
-
In NCBI Virus Search by sequence input form enter NCBI sequence accession sequence in plain text or FASTA format and click Start search.
-
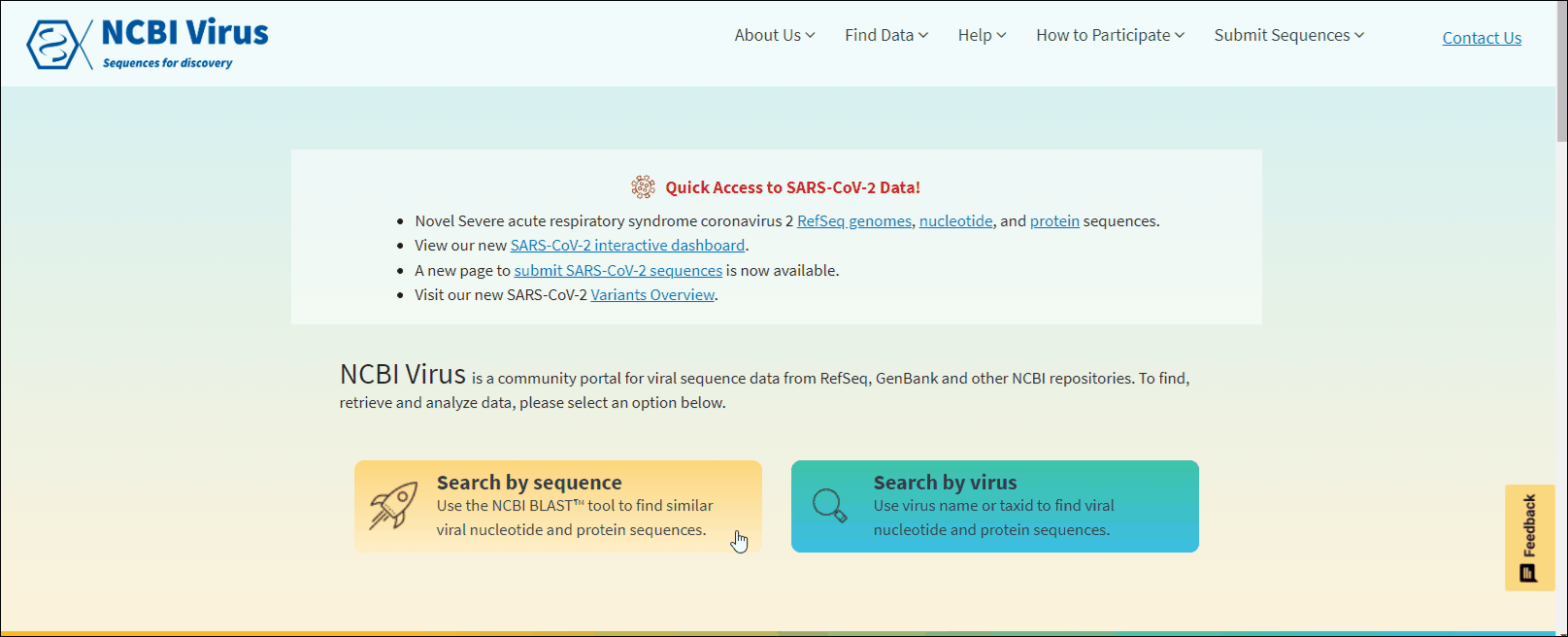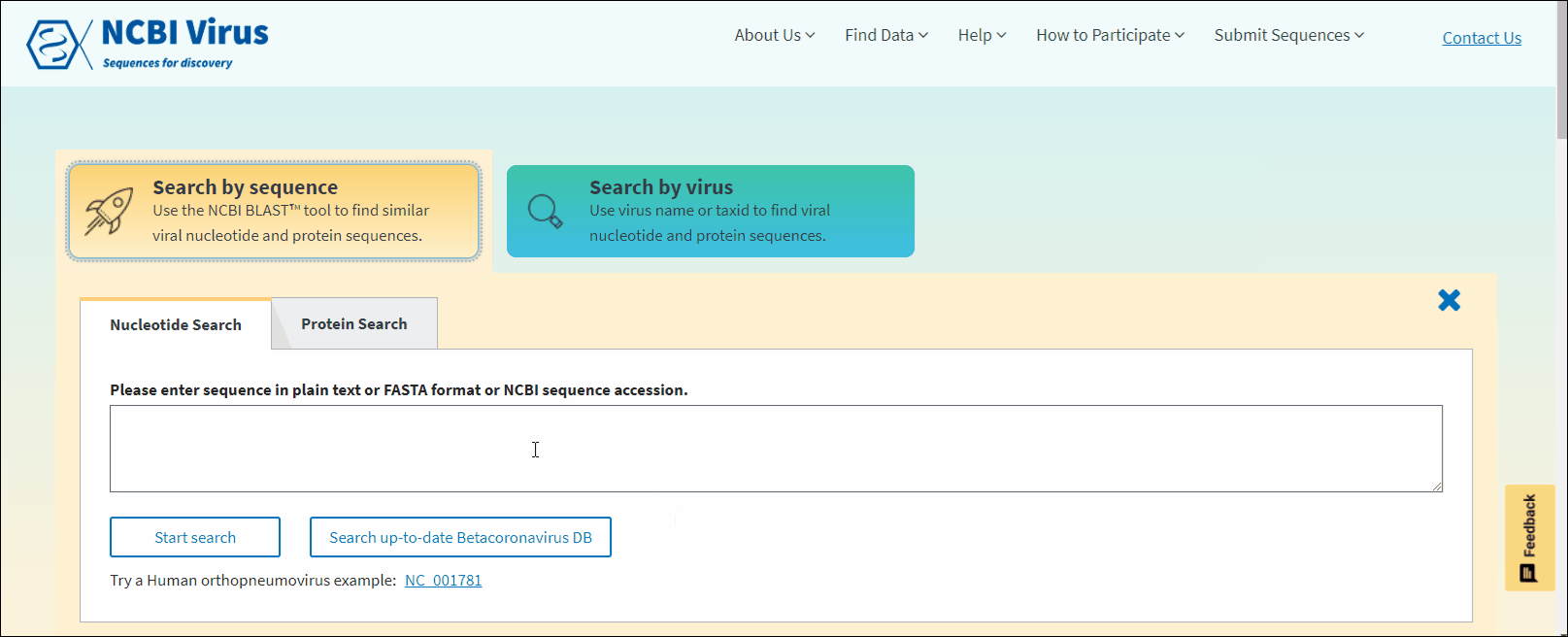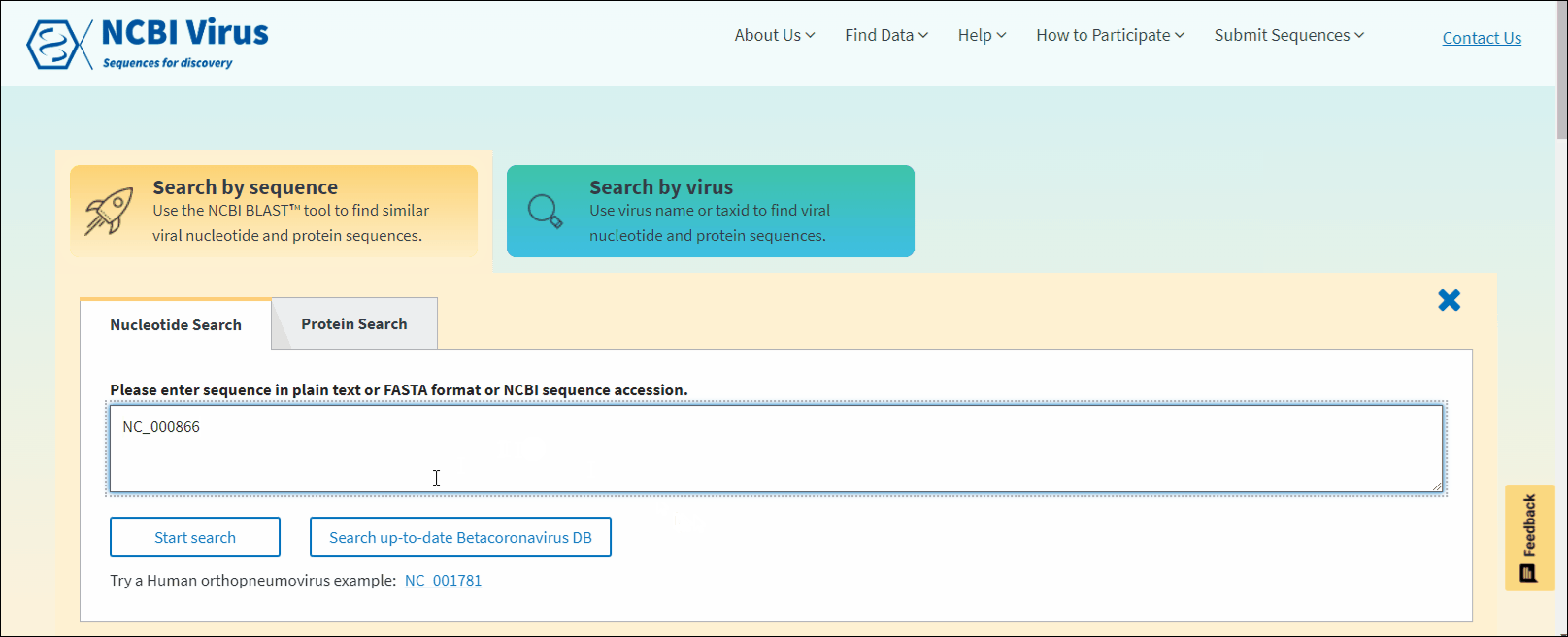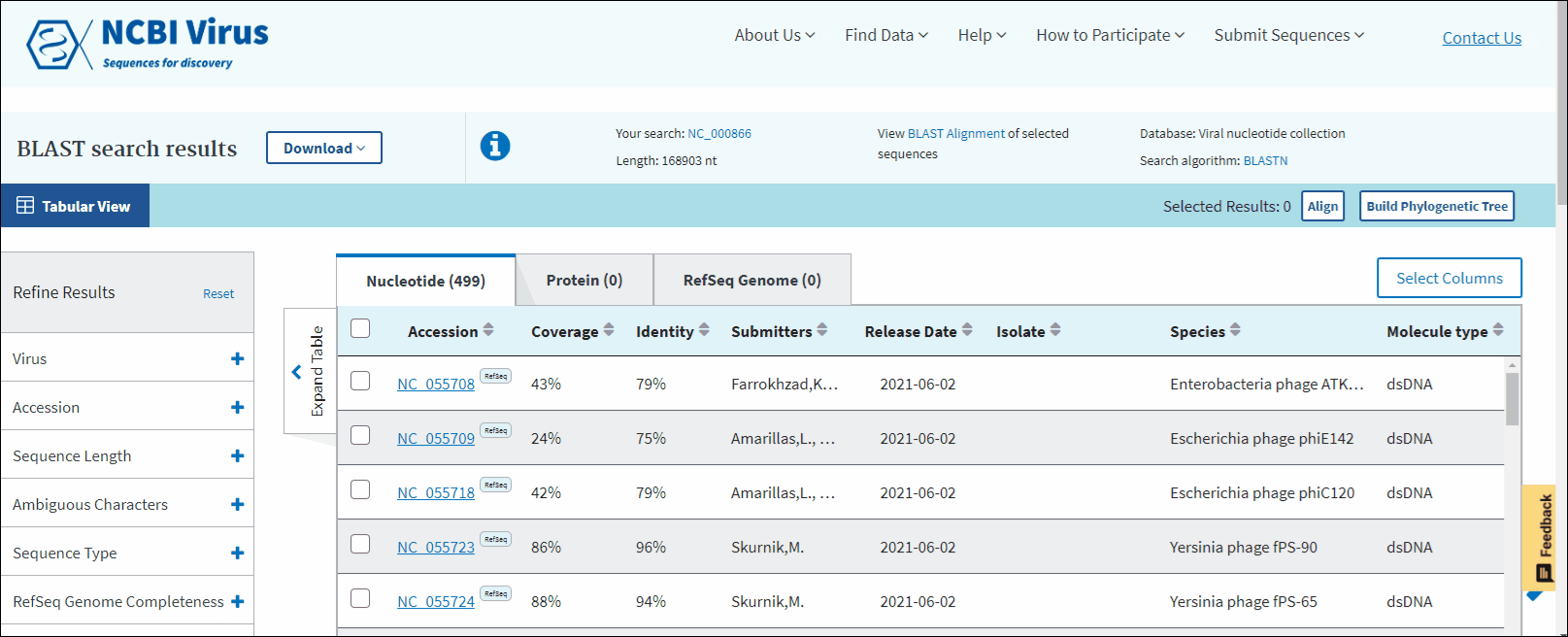
The BLAST search results will open in a new window, presented in a tabulated format (the Results Table).
Compare your sequences to the sequences in up-to-date Betacoronavirus database
To accommodate the SARS-CoV-2 outbreak the Betacoronavirus blast database was created. It is regularly updated and includes all sequences from the genus Betacoronavirus. To search your sequence in Betacoronavirus database using BLAST:
-
Press on the button Search by sequence (or select this option from the Find data navigation tab on the top of the page).
-
Select Nucleotide or Protein tab.
-
In NCBI Virus Search by sequence input form enter NCBI sequence accession sequence in plain text or FASTA format and click Search up-to-date Betacoronavirus DB button.
-
The BLAST search results will open in a separate window in a tabular format (the Results Table).
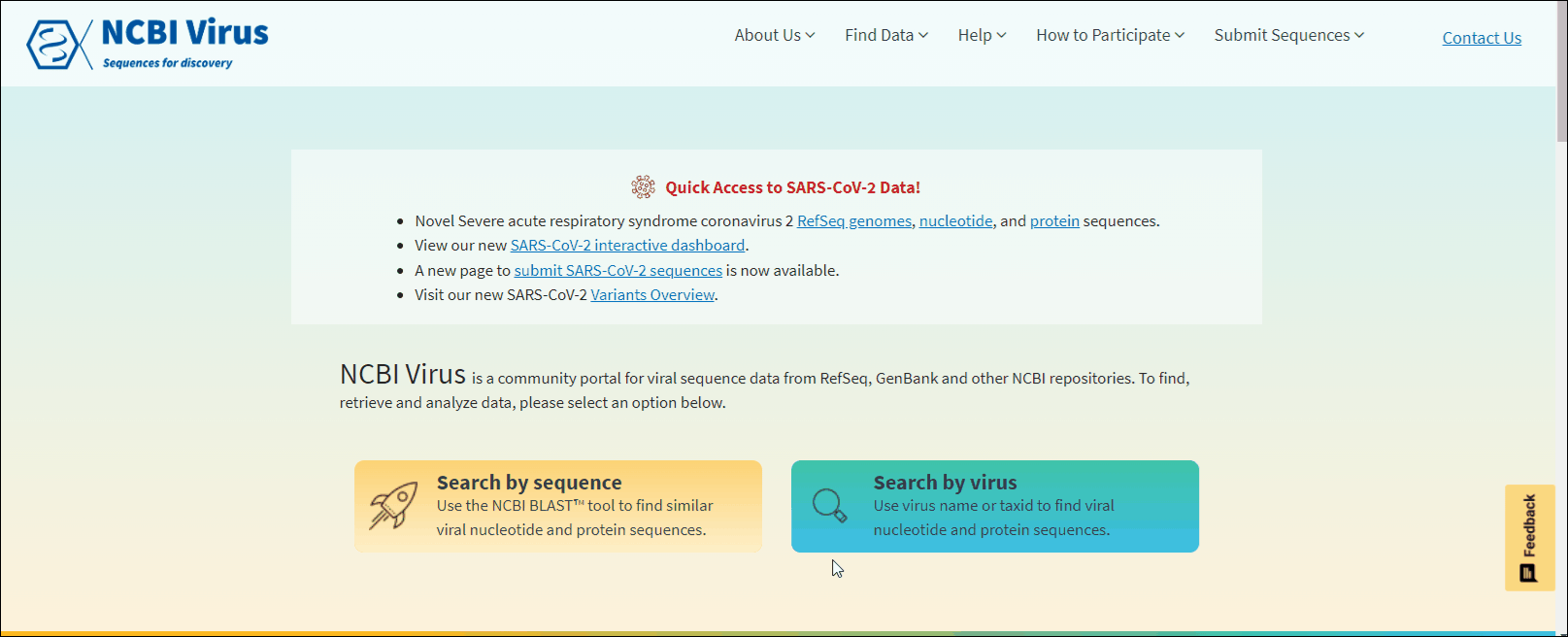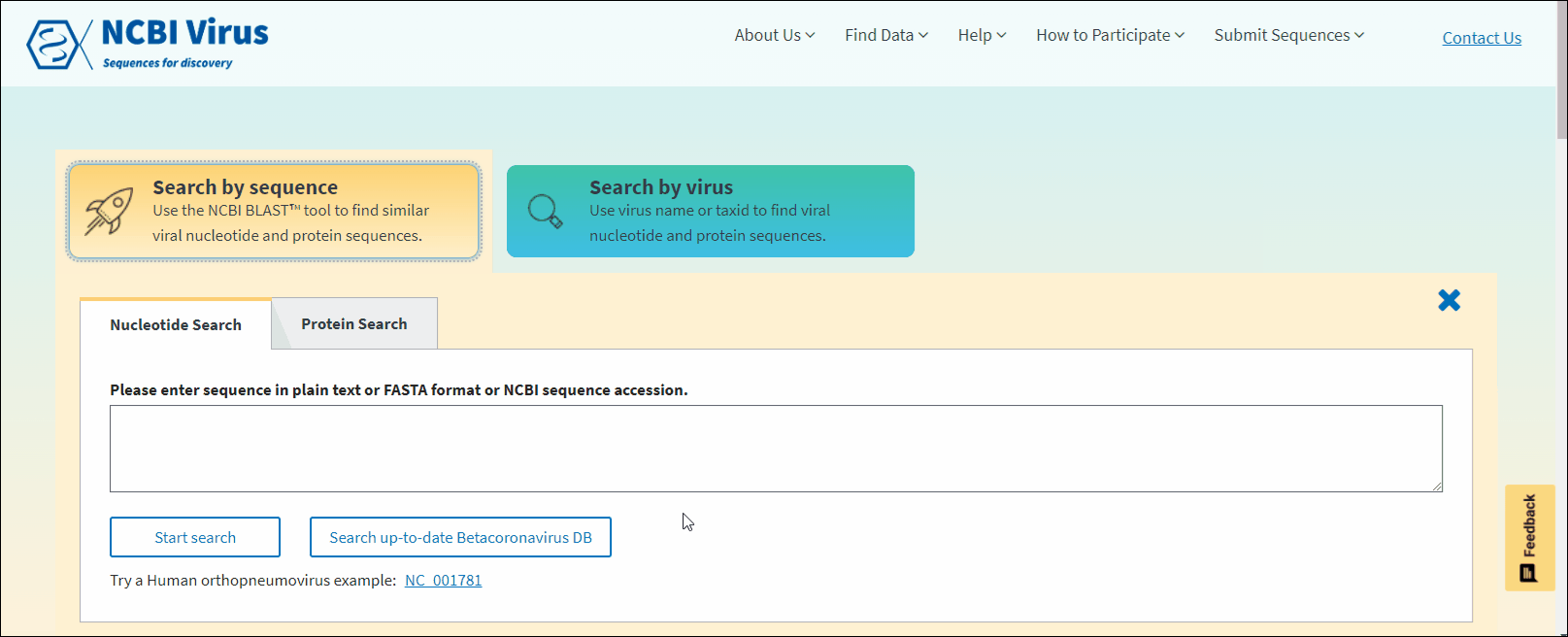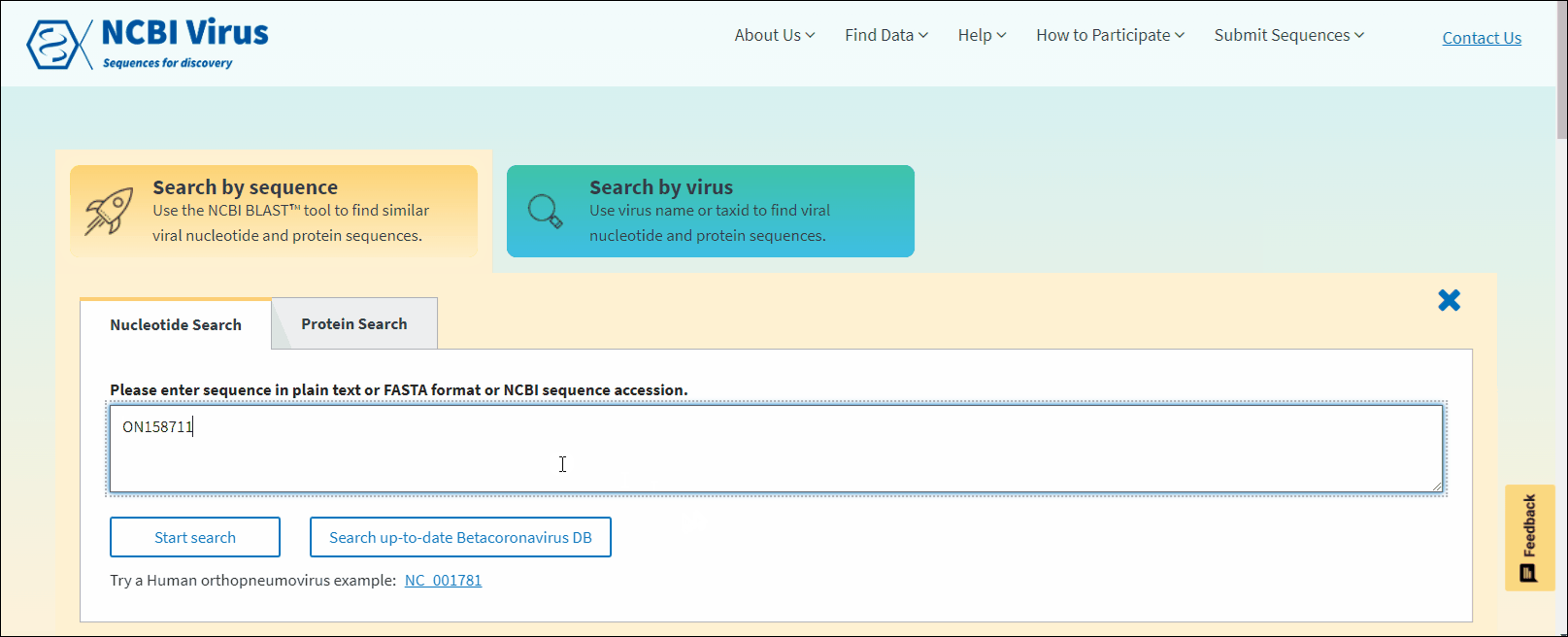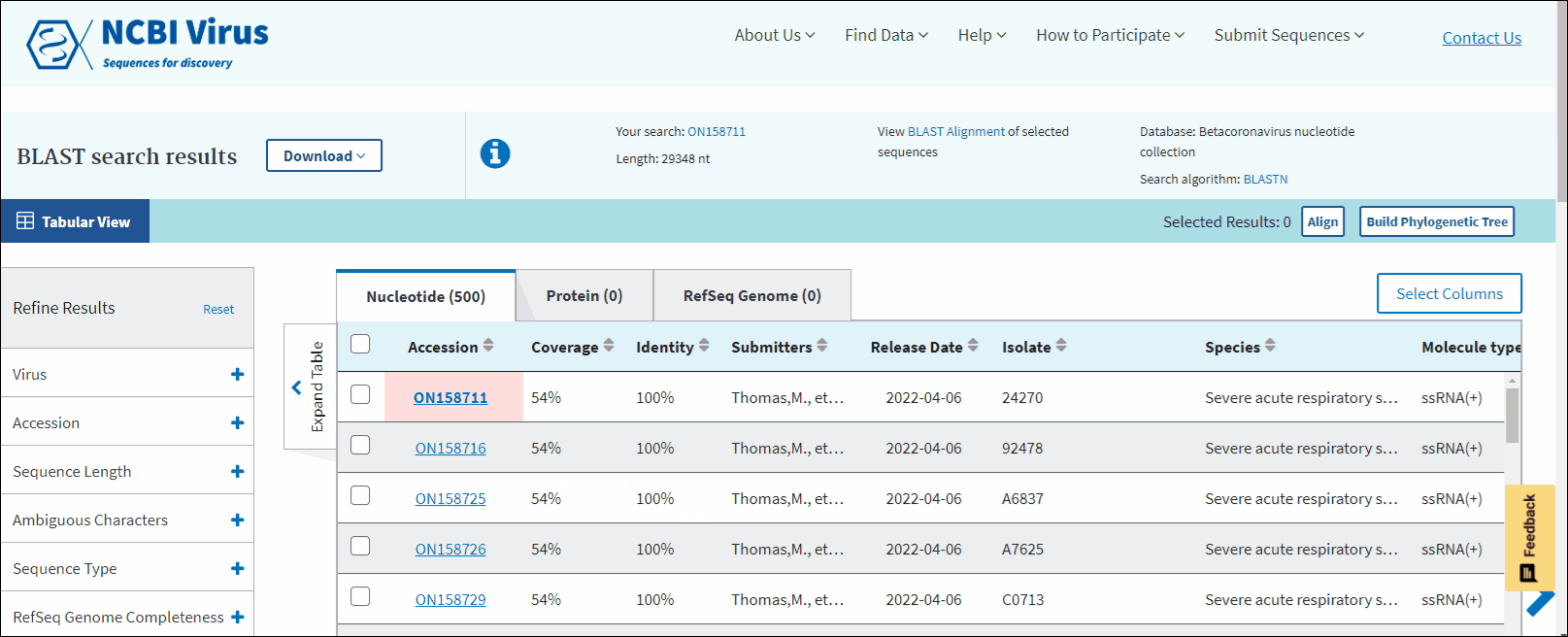
Compare BLAST results in the Results Table
Nucleotide tab allows to perform BLASTN search (using Megablast - optimize for highly similar sequences - search against all NCBI virus nucleotide sequences).
Protein tab allows to perform BLASTP search (search against all NCBI virus protein sequences). Read more about BLAST algorithms on NCBI BLAST help documentation.
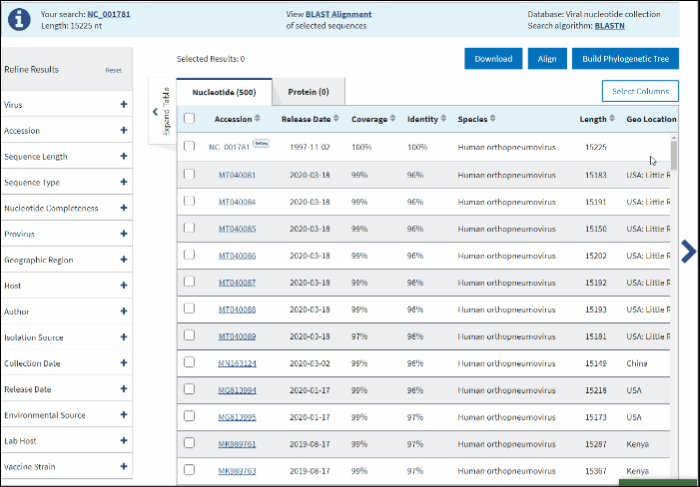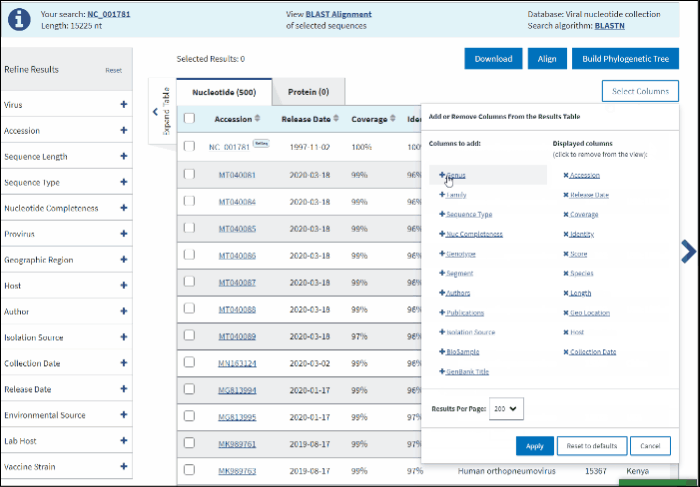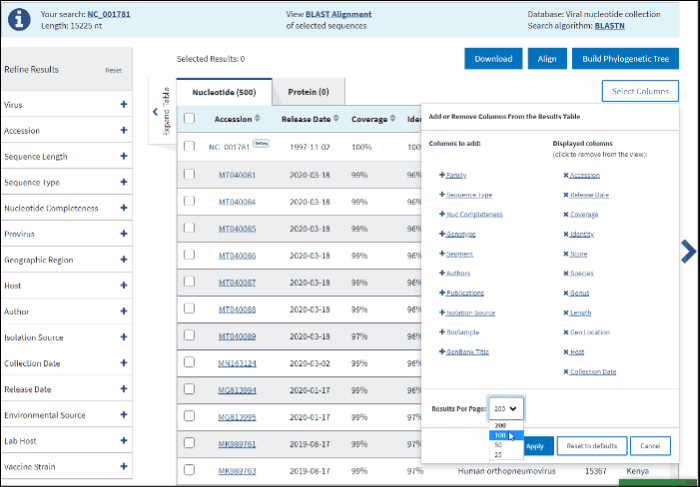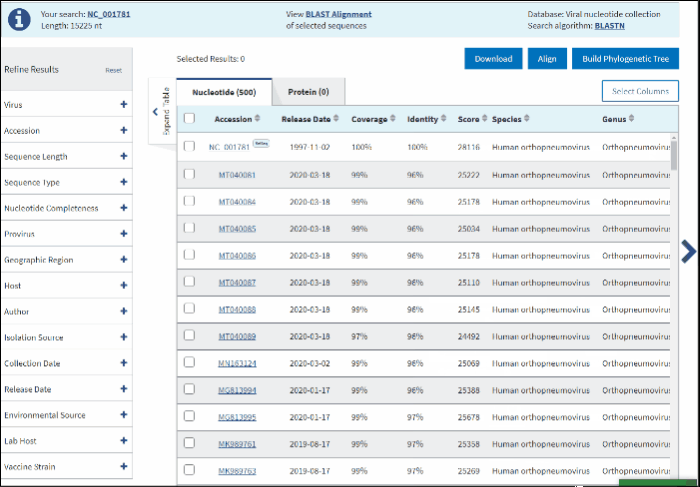
In BLAST search Results Table you can compare search results in tabular display using the following sortable default columns:
-
Accession - the NCBI accession number of the NCBI Virus database sequence. Reference sequence accessions marked with label "RefSeq".
-
Coverage - query coverage.
-
Identity - the highest percent identity of all query-subject alignments.
-
Submitters - authors submitted the sequence. Only first submitter's name is displayed in the column (for example, Baranov,P.V., et al.). To obtain a full list of submitters, click on sequence accession number, this will open the details menu. Click on accession number in the details panel, this will open GenBank Entrez page with all information available for the selected sequence. Alternatively, you can use Download button with CSV format option. The column "Submitters" in the downloaded table will contain the name of all authors submitted each sequence.
-
Release date - the date when sequence was released (publicly appeared) in GenBank or other INSDC databases.
-
Isolate - Individual isolate from which the sequence was obtained, typically an alphanumeric sample ID. Isolate name parsed from "/isolate" field of GenBank record. SARS-CoV-2 sequence isolate name is formatted according to the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) definitions.
-
Species – virus species name.
-
Molecule type - viral nucleic acid type. Molecule type is provided by International Committee on Taxonomy of Viruses (ICTV) in the Master Species List and maintained in the NCBI Taxonomy database. RefSeqs that have "Unknown" molecule type belong to tax groups which were not recognized by the ICTV yet.
-
Length - sequence length.
-
Geo Location - country/region of virus specimen collection. May contain additional geographic information, for example, US state.
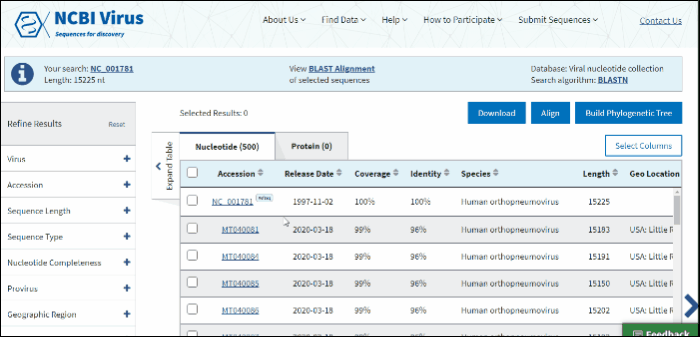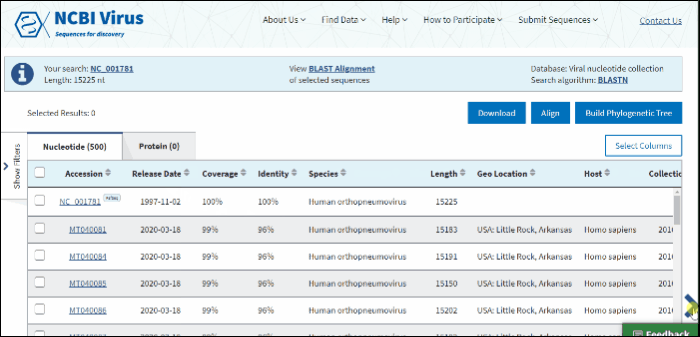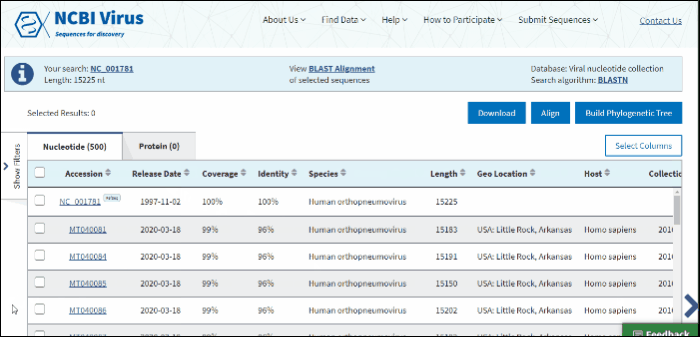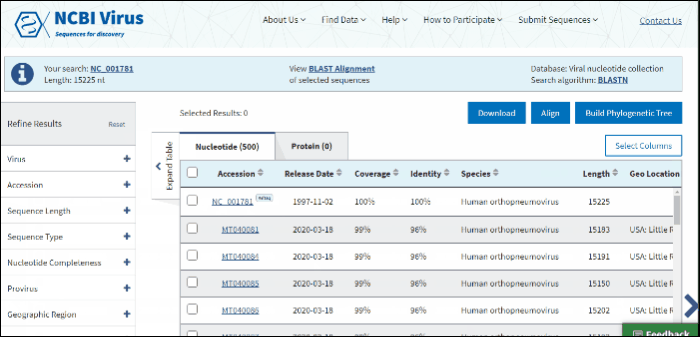
BLAST results can be customized by adding/removing additional columns from the Results Table in Select columns drop-down menu.
Additional columns include:
USA. If the sample was collected in the United States, the column shows the state abbreviation.
Host – virus isolation host (read more about isolation host vocabulary mapping). If isolation host is unknown (/host field of the GenBank record), but laboratory host is present (as indicated in /lab_host field of the GenBank record), the laboratory host will be present in the host column of the Results Table. If both isolation host and laboratory host can be mapped, only isolation host will be presented in the host column of the table.
Collection Date – virus specimen collection date.
-
SRA accession - NCBI Sequence Read Archive (SRA) accession number.
-
Score - the total alignment scores (Total score) from all alignment segments.
-
Genus.
-
Family.
-
Sequence type – complete/partial/proviral/refseq read more about sequence type here.
-
Nuc completeness - nucleotide completeness (note: it is preliminary data, not always accurate).
-
Genotype.
-
Segment – segment name in case of segmented viruses.
-
Publications - links to the associated with sequences publications in PubMed.
-
Country - country of specimen collection (only country, no any additional information).
-
Isolation source – sequence isolation source read more about isolation source here.
-
BioSample – NCBI BioSample accession number.
-
BioProject – NCBI BioProject accession number.
-
GenBank title.
The default number of rows displayed in the Results Table is 200. You can change the number of table rows by selecting number results per page (200, 100, 50 or 25) in Select Columns menu.
View BLAST Alignment of selected sequences
To compare search results in pair-wise alignment:
-
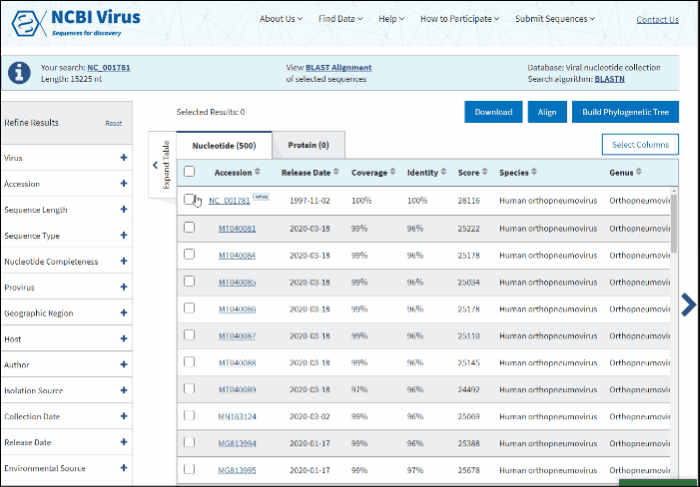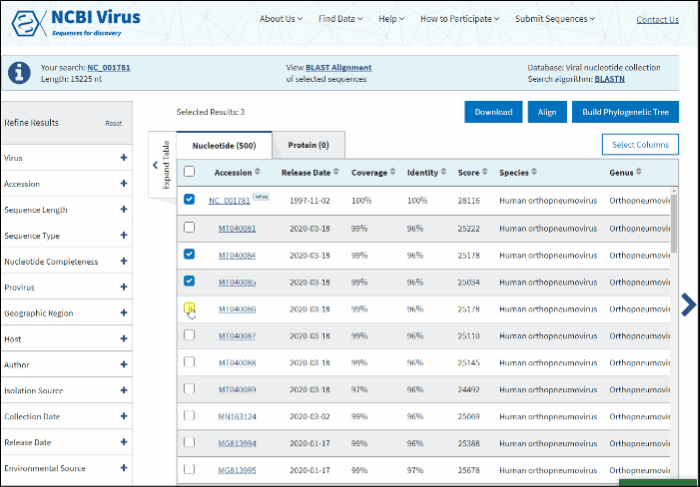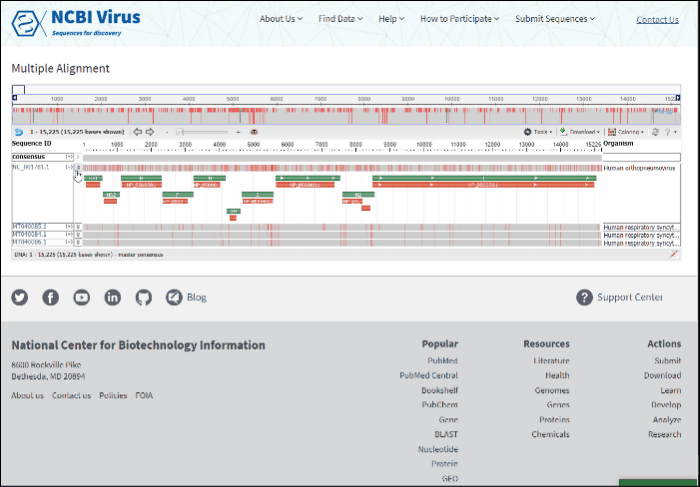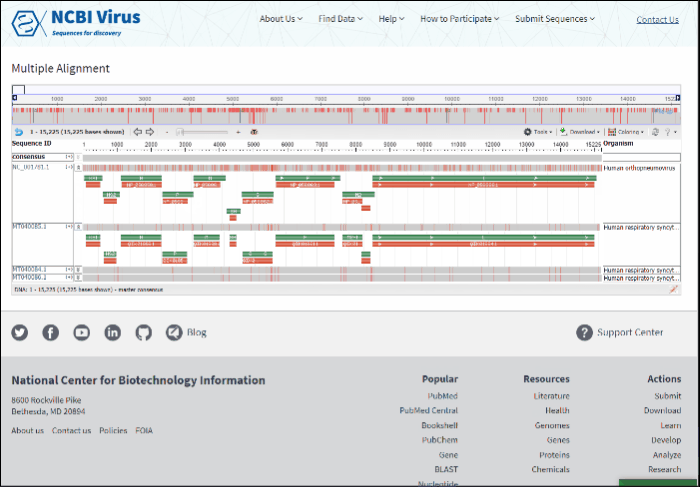
Select sequences to display.
-
Click on View BLAST Alignment of selected sequences link displayed in the center of the Info panel located above the Results Table.
The new page will show a graphical view of pairwise alignments between selected BLAST results and the query, along with a feature map (if available) of the query at the top of the view.
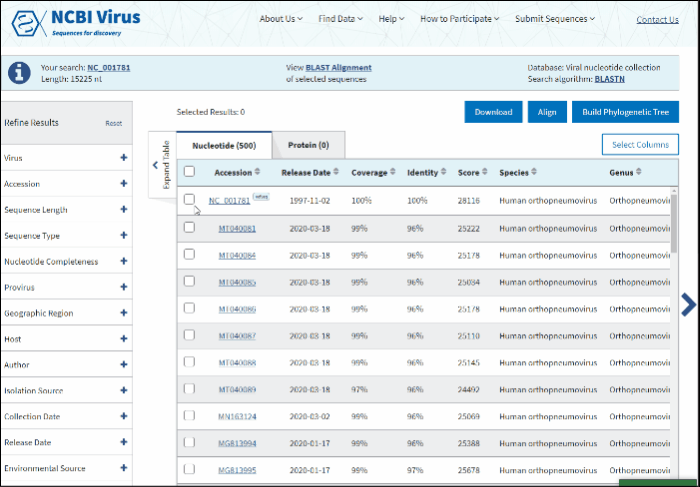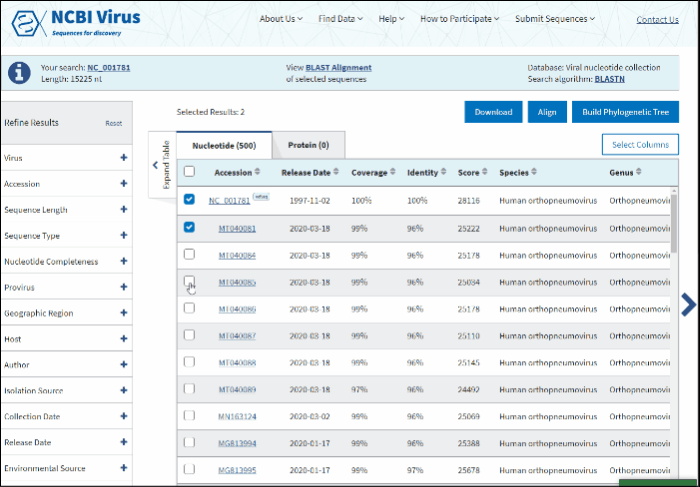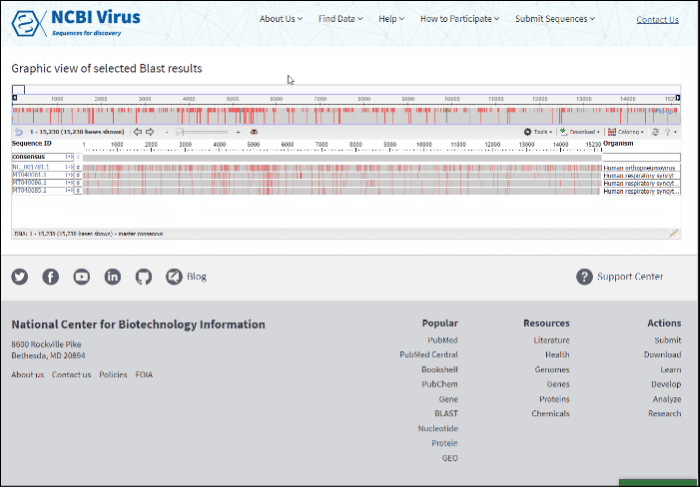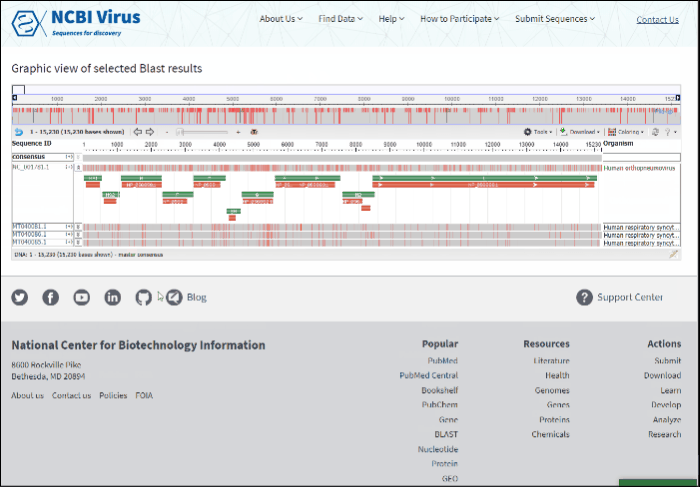
Read more how to use alignment viewer please refer to NCBI Multiple Sequence Alignment Viewer documentation.
Build multiple sequence alignment of selected BLAST results
To build multiple sequences alignment based on selected BLAST results:
-
Select sequences that you want to align.
-
Press the button Align on the right above the Results Table.
Multiple sequence alignment will open at the new page. Multiple sequence alignments calculated using MUSCLE.
Read more how to use alignment viewer please refer to NCBI Multiple Sequence Alignment Viewer documentation.
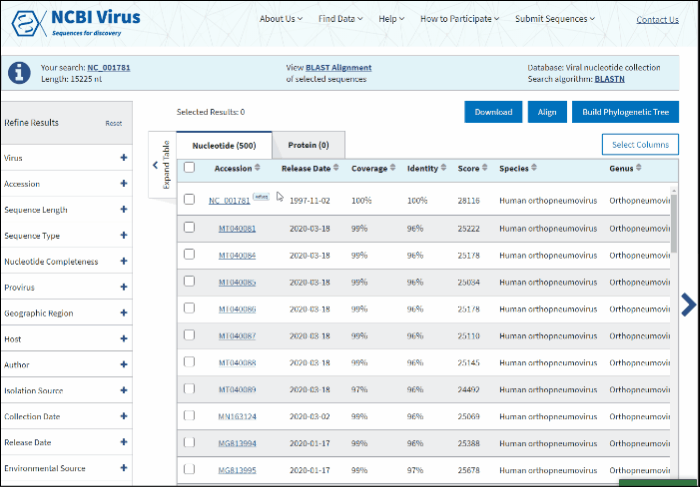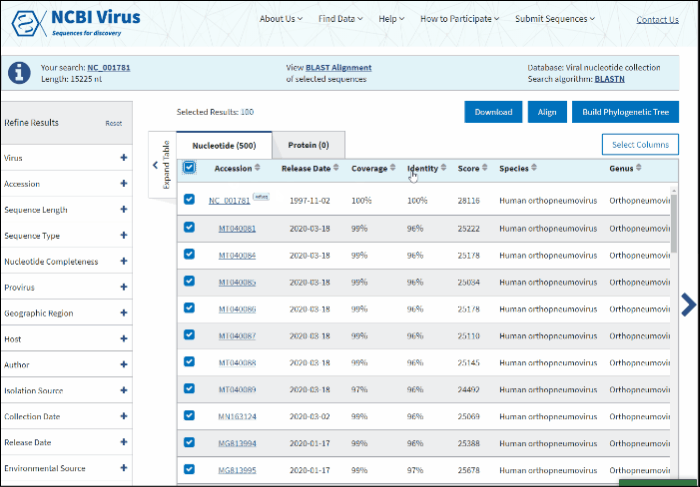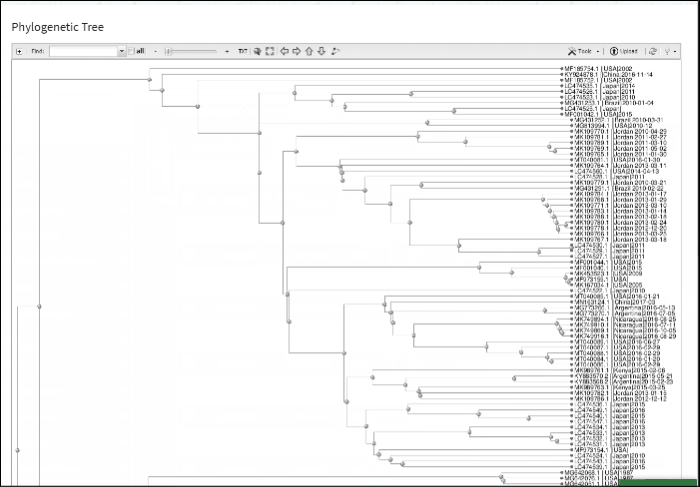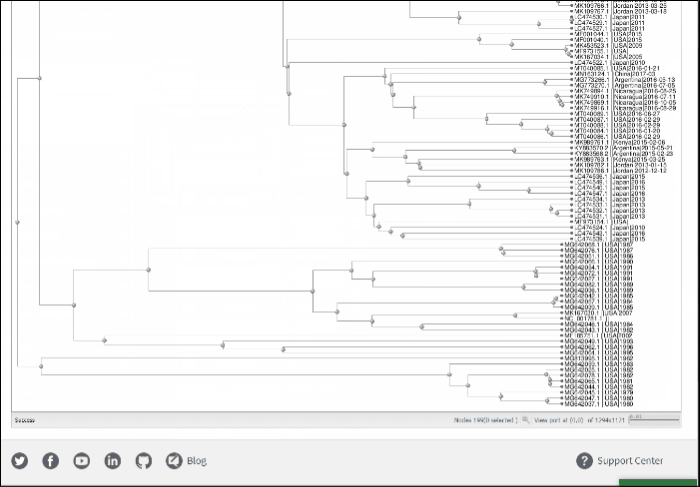
Build phylogenetic tree of selected BLAST results
To build a phylogenetic tree to see the relationships of selected sequences:
-
Select sequences to display.
-
Press the button labeled Build Phylogenetic Tree on the right above the Results Table.
The tree will be calculated and available in tree viewer on a separate page.
For more about Tree Viewer and how to use it, please refer to NCBI Tree Viewer help documentation located here.
Refine tabular BLAST results via filters:
1. Virus name or taxonomy
To Restrict search results to the particular virus group:
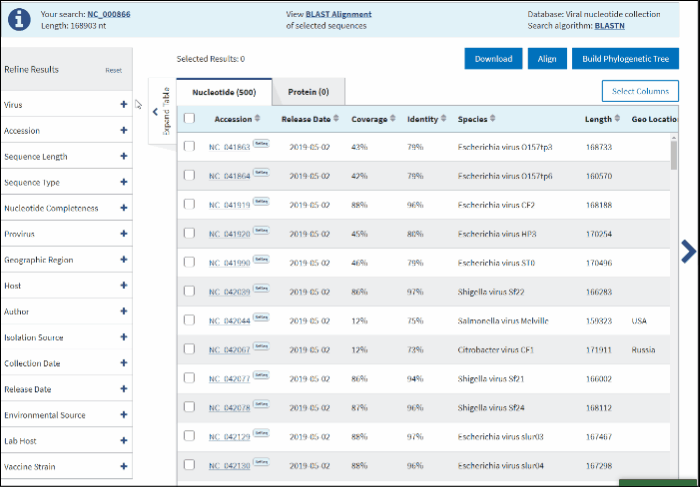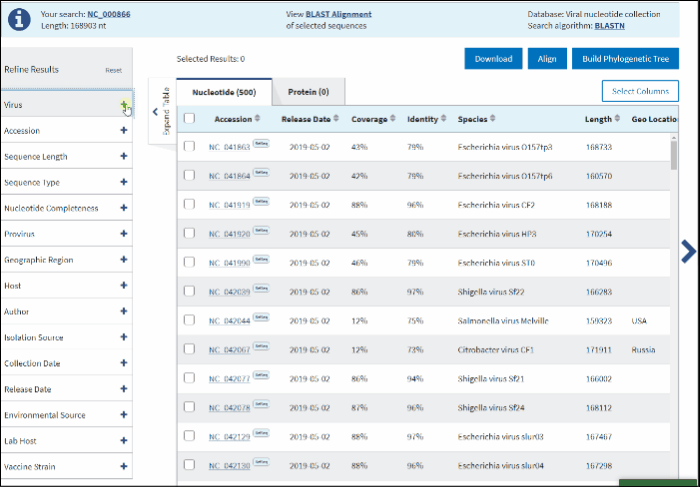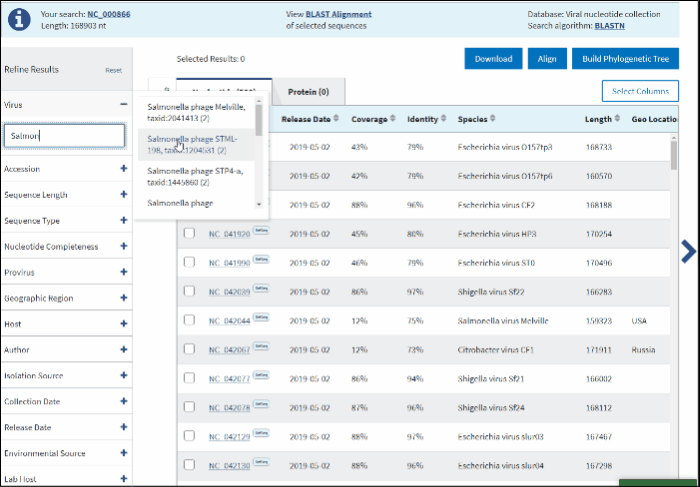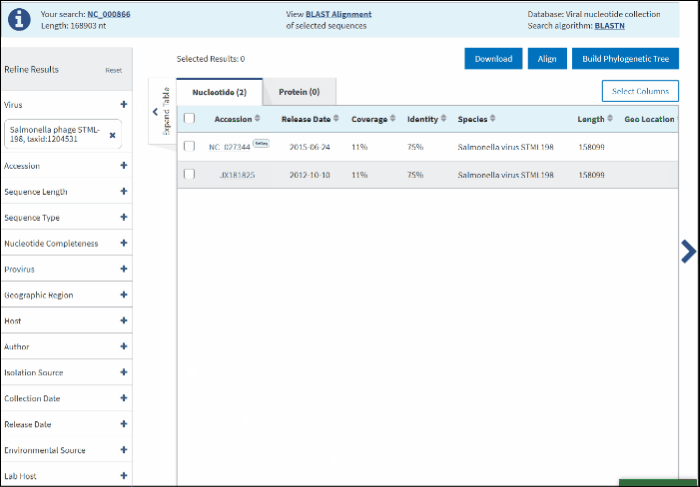
On BLAST result page in Refine Results panel (left upper corner) click on Virus.
In the text box paste or start typing a single virus taxonomy name, or taxid (only 5 top taxa will be shown).
Select your taxid (NCBI taxonomy database ID) from the fly-out menu.
The filtered results will be presented in the Results Table with the following 5 default sortable columns: accession, coverage, identity, species, country, host, collection date. Additional columns to display connected metadata can be added via the Customize Table menu. The query sequence will be highlighted in the first row of the table.
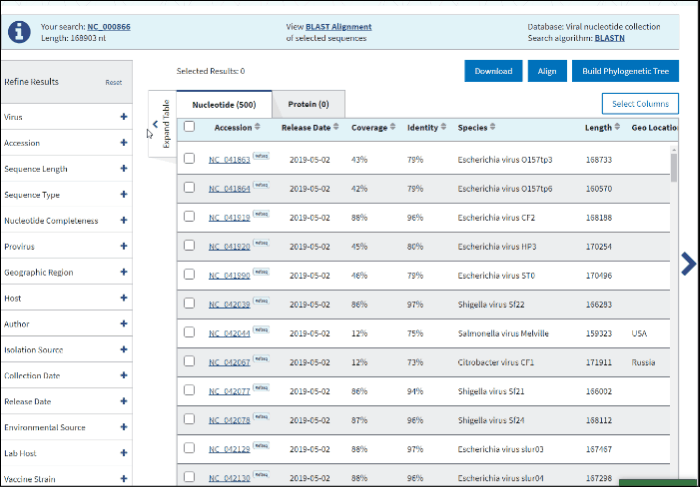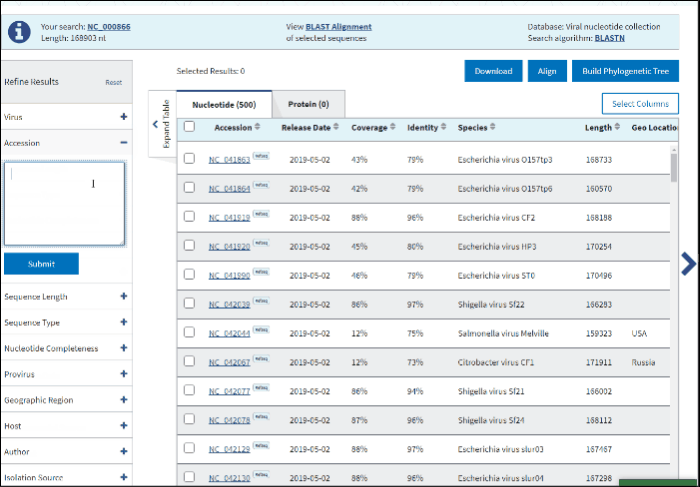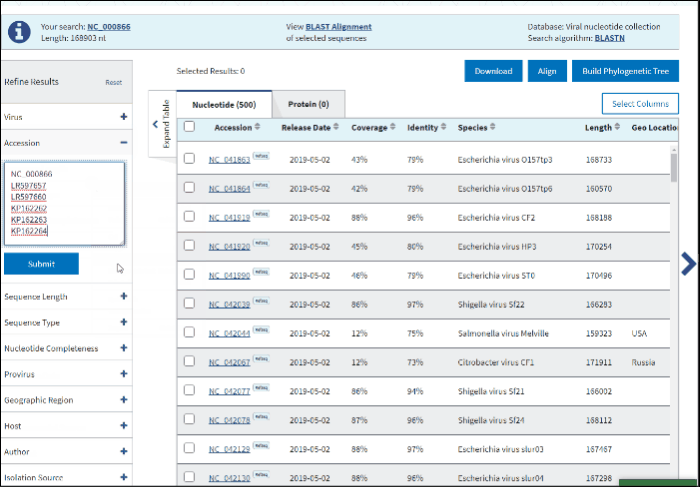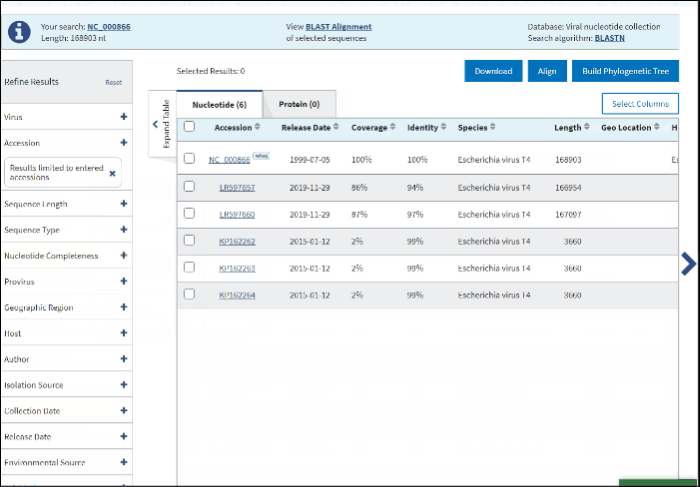
2. Accession
You can search for the particular accessions in the Results Table by entering them in the search form under the Accession filter. The results on the table will be limited to the entered accession numbers.
3. Sequence length
To restrict your results to the particular sequence length, enter the minimum and maximum length in nucleotides (for nucleotide search) or amino acids (for protein search).
4. Ambiguous Characters
Allows to set the desired maximum number of ambiguous characters (N's in nucleotide or X's in protein) in each sequence on the Results Table.
5. Sequence type
All sequences (Nucleotide or Protein) available in the NCBI Virus resource can be filtered based on following sequence types - GenBank and RefSeq.
GenBank sequences include all sequences available in GenBank, except RefSeqs.
Refseq filtered nucleotide sequences include all reference sequences for the selected virus. Note, that few RefSeqs are partial genomes, based on the International Committee on Taxonomy of Viruses (ICTV) proposal.
6. RefSeq genome completeness
Complete or partial RefSeq genomes - filter for all complete (or partial) genomes, reference records (RefSeqs), and proteins form these RefSeqs. In case of segmented viruses complete genomes contain all genome segments. Most of RefSeq records are complete, but few RefSeqs are partial, based on International Commitee on Taxonomy of Viruses (ICTV) proposal.
7. Nucleotide completeness
Complete nucleotide sequences - filter for all NCBI viral nucleotide sequences, where GenBank ASN.1 format contains the following descriptors: descr/molinfo/completeness=complete or there is a word 'complete' present in the record’s definition line (defline). It also includes complete reference records (RefSeqs).
Partial nucleotide sequence – filter for sequences that are not complete according to the definition above.
If Protein tab selected and complete nucleotide sequence type filter applied, results will include all proteins from complete genomes or individual complete segments in case of segmented viruses.
8. Isolate
Isolate - individual isolate from which the sequence was obtained, typically an alphanumeric sample ID. Isolate name parsed from "/isolate" field of GenBank record. SARS-CoV-2 sequence isolate name is formatted according to the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) definitions.
9. Proteins
Protein name parsed from "/product=" field of GenBank nucleotide and protein records
10. Provirus
Provirus sequences - filter for sequences that have "/proviral" source qualifier in the GenBank record.
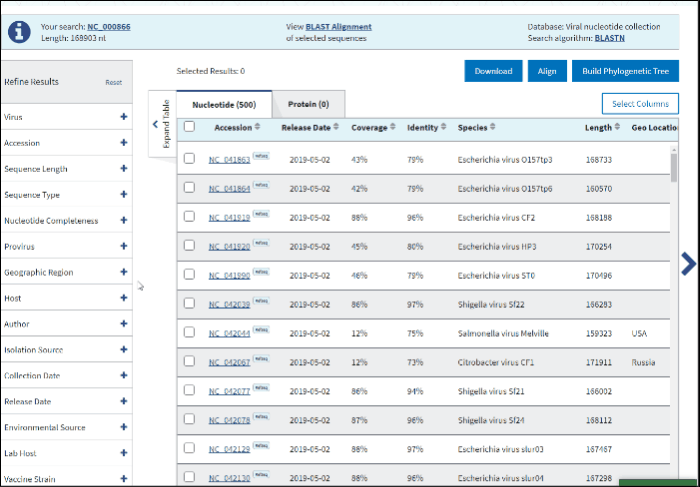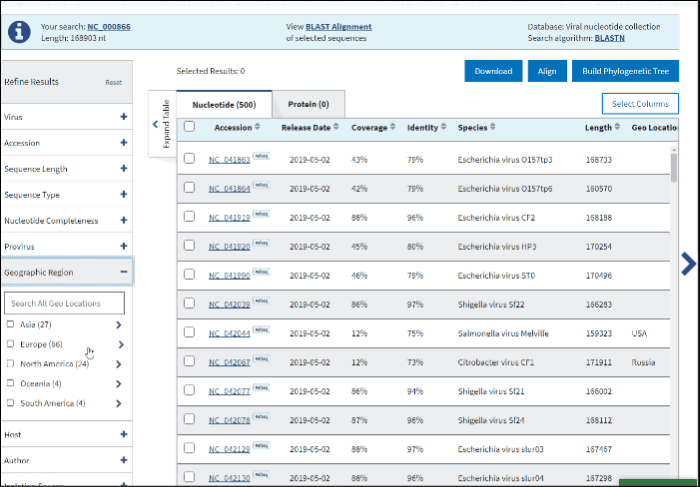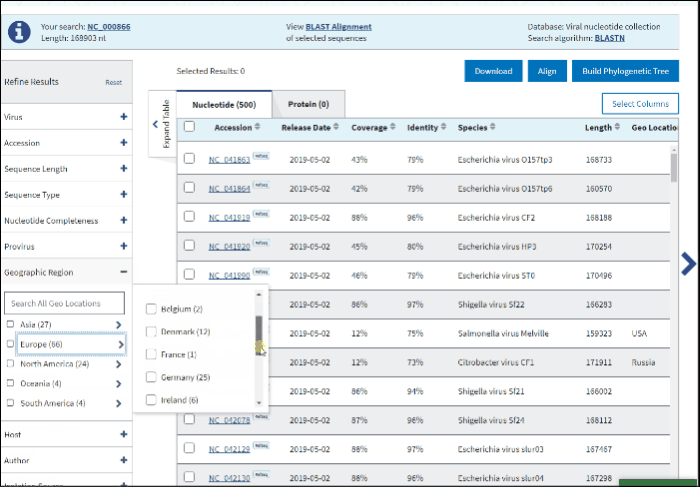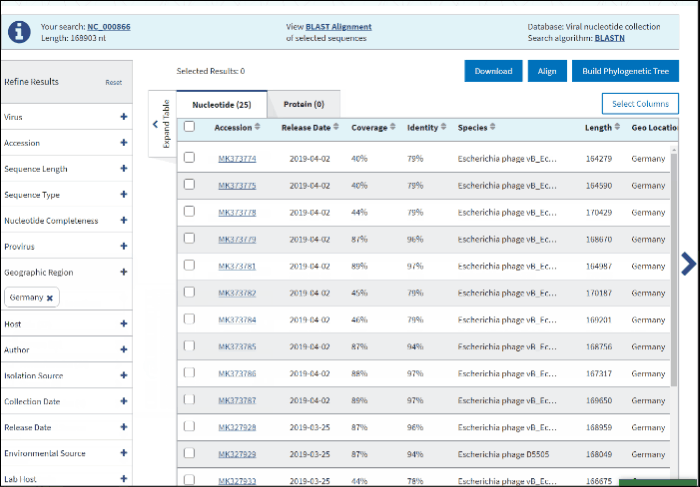
11. Geographic region
The Geographic region filter allows you to type your country of interest in the text box or select the continent(s) of interest. Selecting a continent also selects all the countries within that continent automatically.
Clicking on the arrow next to a continent's name opens a secondary selection menu to (un)select the country(s) belonging to the continent of interest. The selected countries are listed below the continent name.
If an entire continent is selected, the continent's name will be shown in a pillbox below, indicating that all countries for the continent are selected. If at least one country is selected, the corresponding continent is no longer displayed and instead, a pillbox for each selected country is shown below the associated continent. Each continent’s behavior is independent of the other continents.
Selection can be deselected by clicking on the pillboxes, and multiple concurrent selections are supported.
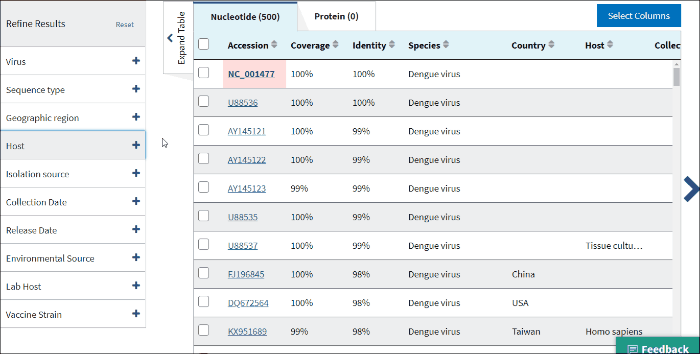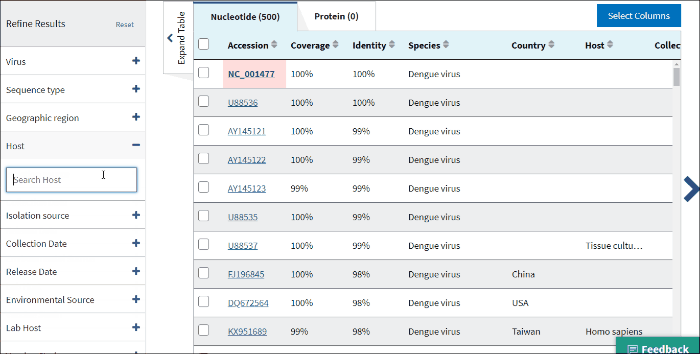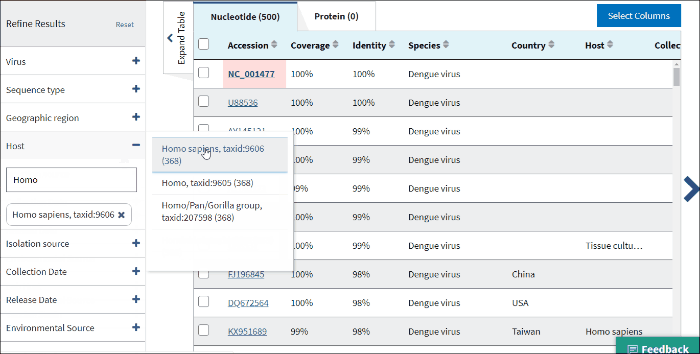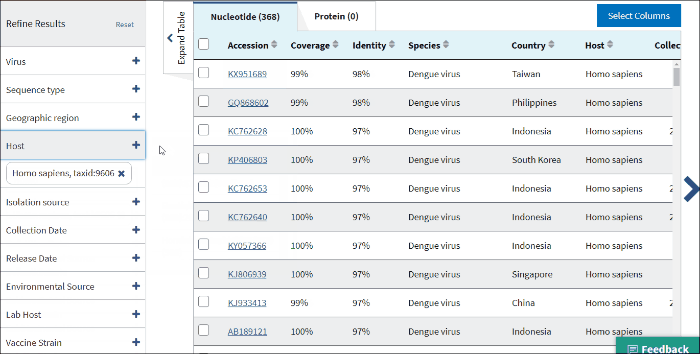
12. Isolation host or taxonomy
Enter a host name or taxid to the text box and several host terms will be suggested (only 20 top taxids will be shown). Select the desired host term and hit Enter. The results will be restricted to sequences in the database with the indicated host term. Multiple hosts can be filtered on simultaneously by adding additional host terms to the filter.
The terms for isolation host are parsed from the source/host field in a sequence's GenBank record. Parsed terms are mapped to a standardized vocabulary, which was derived by curators by aggregating the variety of terms in GenBank files. Common mis-spellings are also included in this mapping strategy. For example, "Accipter cooperii" is mapped to "Accipiter cooperii".
The terms for isolation hosts are displayed in the host column of the Results Table. In case if the isolation source is unknown, but laboratory host is present (as indicated in /lab_host field of the GenBank record), the laboratory host will be present in the host column of the Results Table. If both isolation host and laboratory host can be mapped, only isolation host will be presented in the table (host column).
13. Submitters
To search for sequences submitted by a particular author(s) enter the author's last names with or without initials.
The following formats are supported:
Chiang,T.Y. Forsyth,K.A. Knittig,L.C. Lim,O.P. Chiang,T.Y., Forsyth,K.A., Knittig,L.C., Lim,O.P. Chiang Forsyth Knittig Lim Chiang, Forsyth, Knittig, Lim
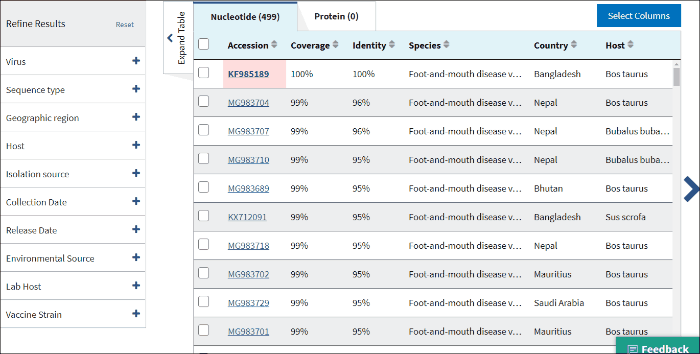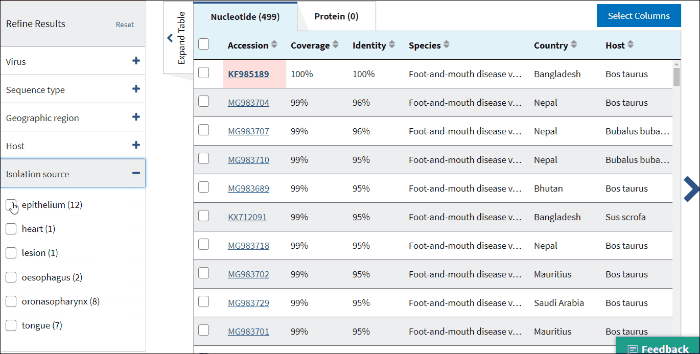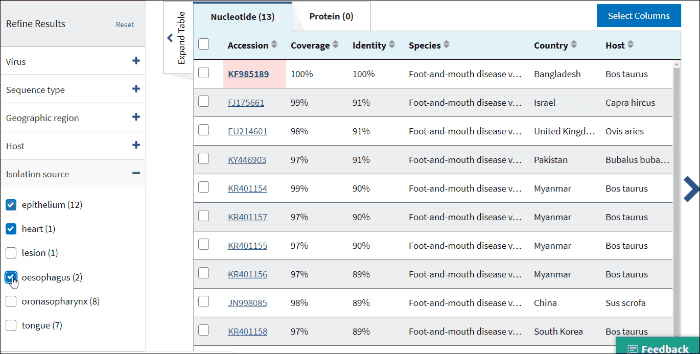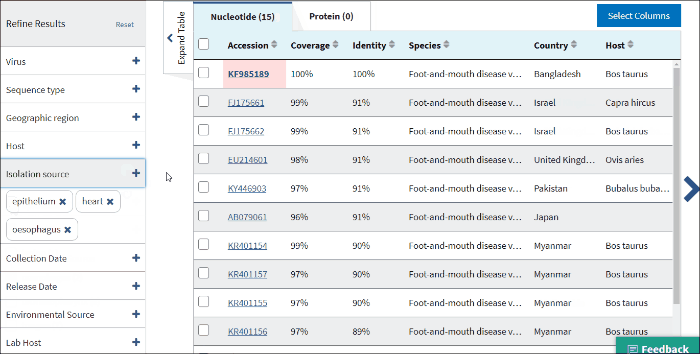
14. Isolation source
The terms for isolation source are parsed from the isolation source field in a sequence's GenBank record. Examples of parsed terms are serum and plasma, which are all mapped to the standardized vocabulary term blood.
Common mis-spelling as well as regional spelling differences are included in the mapping strategy. Multiple terms can be selected.
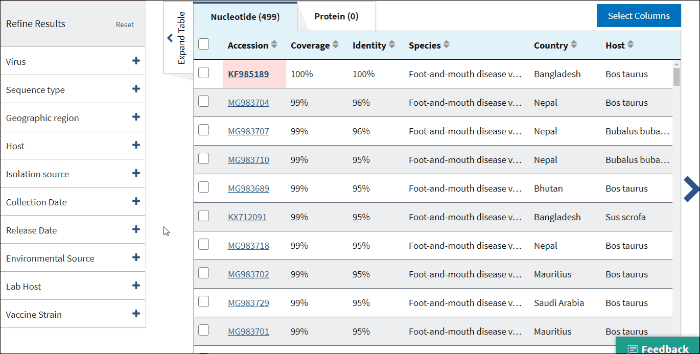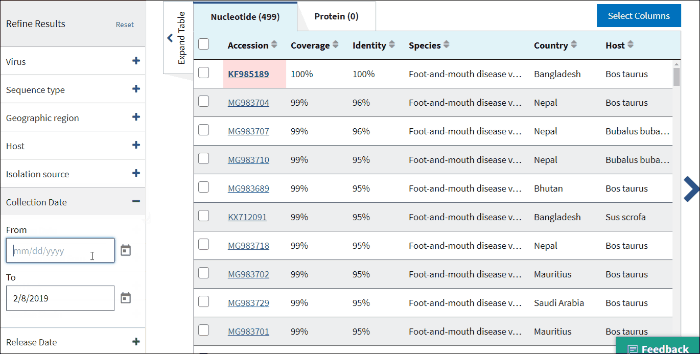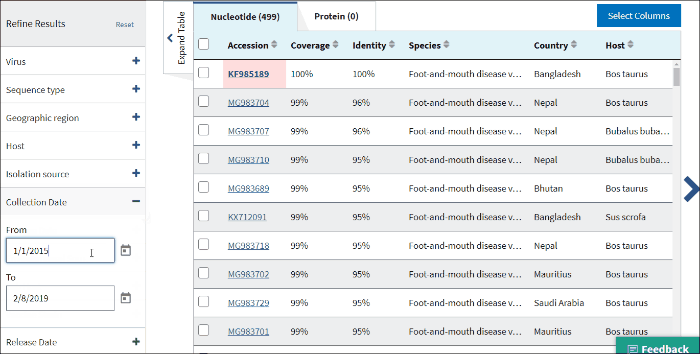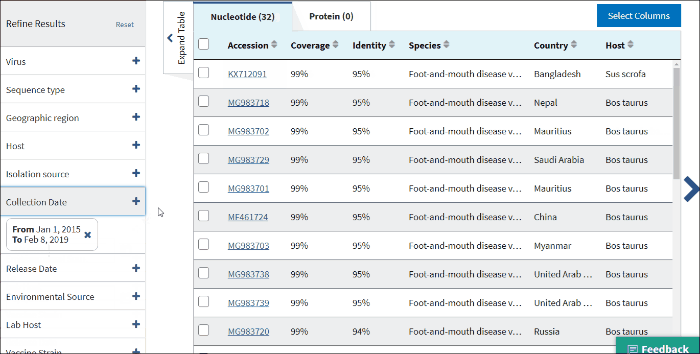
15. Sample collection date
Collection date (From, To) - is the collection date for the sample from which the sequence was derived.
By default, the To: date is set to the current date.
Use mm/dd/yyyy or yyyy formats or click on the calendar icon and select dates.
16. Sequence release date
Release date (From, To) – the date when sequence was released (publicly appeared) in GenBank or another INSDC database.
By default, the To: date is set to the current date.
Use mm/dd/yyyy or yyyy formats or click on the calendar icon and select dates.
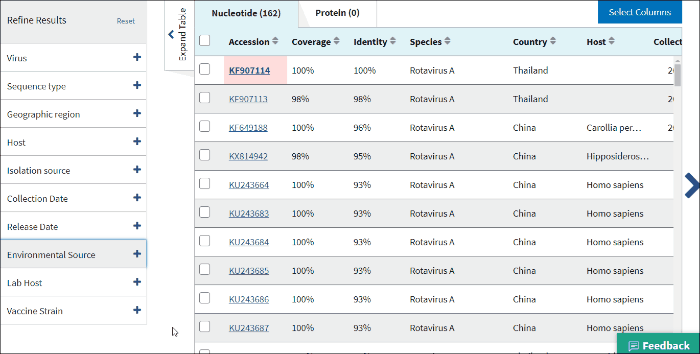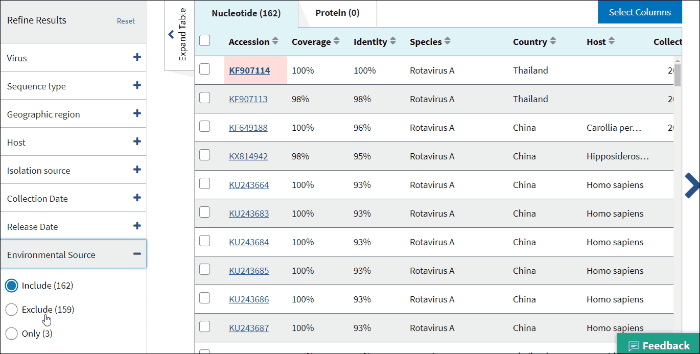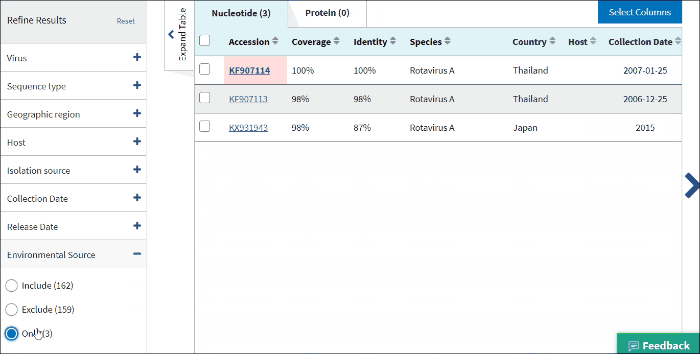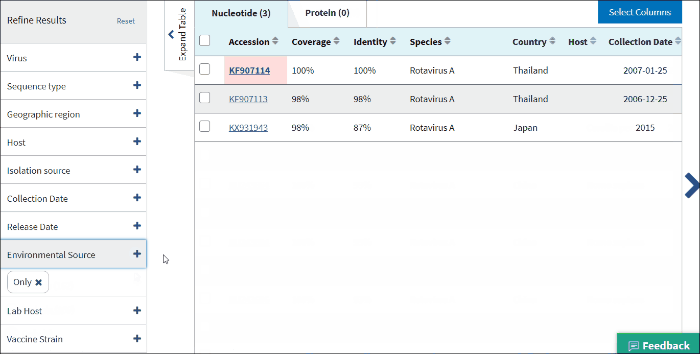
17. Environmental sourse
Environmental source filter allows to select virus sequences isolated from the environmental sources. Generally, environmental isolates are identified by searching for key terms, such as sewage or ocean water from /isolation_source and /note fields of GenBank records when /host field is empty.
Select Include - to include all sequences isolated from environmental sources to the Results Table.
Select Exclude - to exclude all sequences isolated from environmental sources to the Results Table.
Select Only - to view only sequences isolated from environmental sources.
14. Laboratory samples
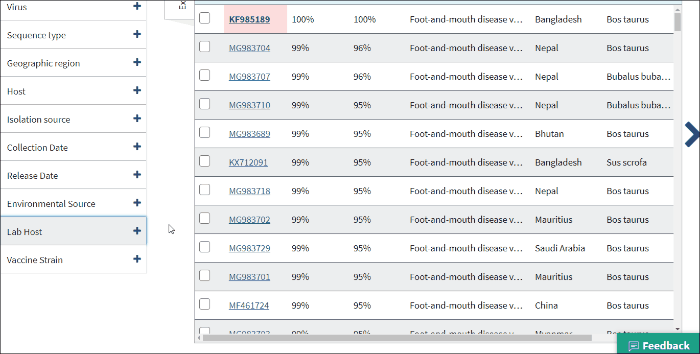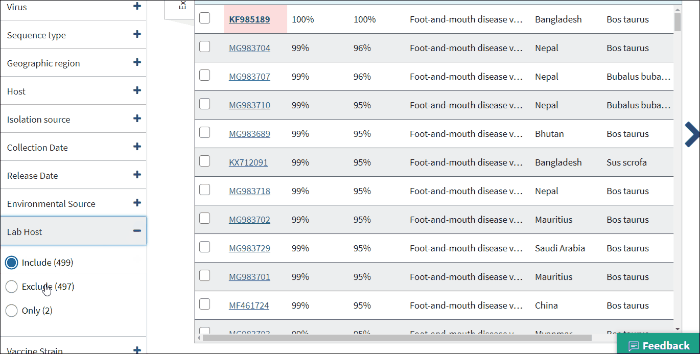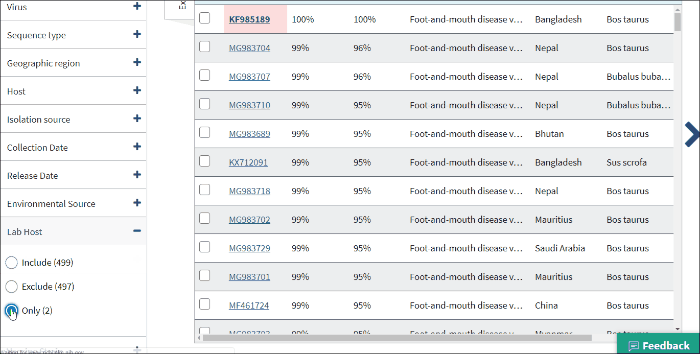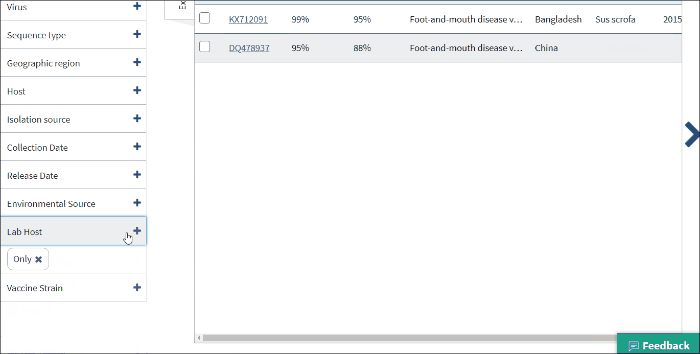
Lab host filter allows to view laboratory isolated virus sequences. Lab host identified by searching lab host name in /lab_host field of GenBank record. Additionally (only for bacteriophages) if /host and /lab_host fields are empty, lab host identified by parsing lab host name from bacteriophage organism name of GenBank record.
Select Include - to include all laboratory isolated virus sequences to the Results Table.
Select Exclude - to exclude all laboratory isolated virus sequences to the Results Table.
Select Only - to view only laboratory isolated virus sequences.
Note: lab host name can be viewed in the result table (in host column) only in cases when the isolation host cannot be identified (/host field of GenBank record is empty).
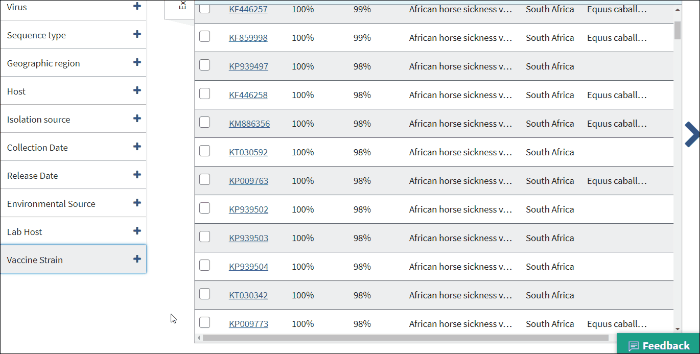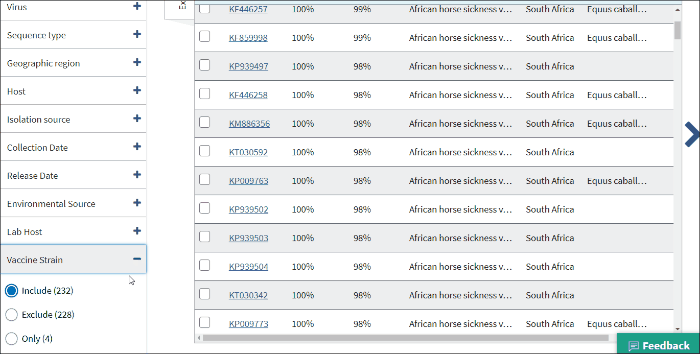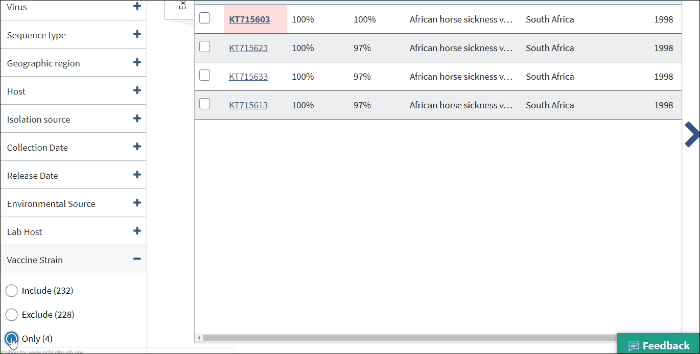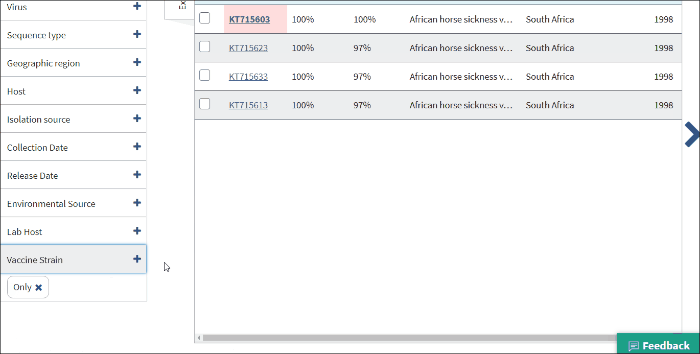
15. Vaccine strain
Vaccine strain filter allows to find virus vaccine strain sequences. Vaccine strains identified by searching vaccine strain terms in /isolation_source, /note, /host and definition line of GenBank record.
Select Include - to include all virus vaccine strain sequences to the Results Table.
Select Exclude - to exclude all virus vaccine strain sequences to the Results Table.
Select Only - to view only virus vaccine strain sequences.
Search for sequences by virus name or taxonomy group
Find your virus sequence(s)
Option 1:
Select Search by virus drop-down option from navigation menu Find Data tab on any of NCBI Virus pages. This will open the selection menu.
Start typing in the text box, then select your taxid (NCBI taxonomy database ID). To select all viral sequences, enter and then select the term viruses.
The results will be shown in the table.
Note: Please view a list of all viral taxonomy terms using the NCBI taxonomy pages.
Option 2:
Click on button Search by virus located in the central part of NCBI virus home page.
Start typing in the text box, then select your taxid (NCBI taxonomy database ID).
This will open the tabular interface with sequences from the selected taxonomy group.
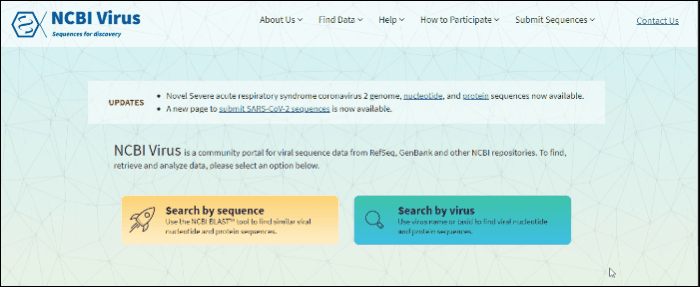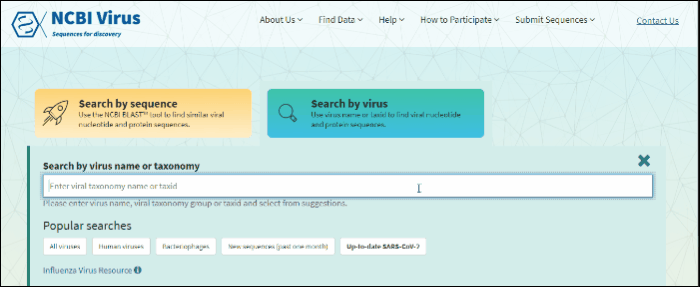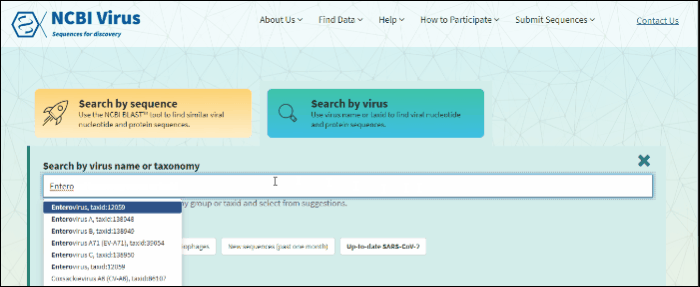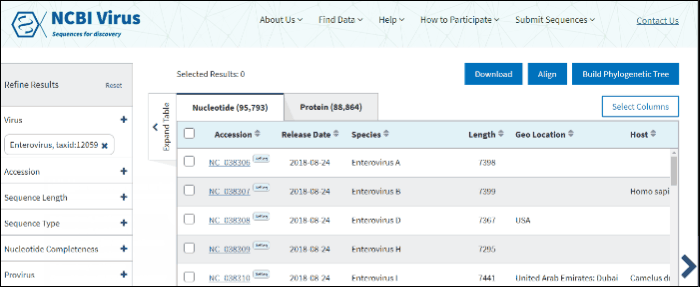
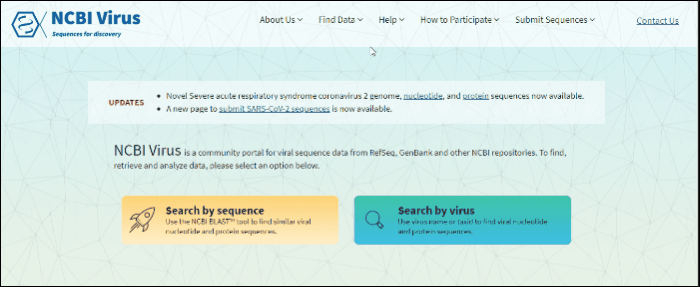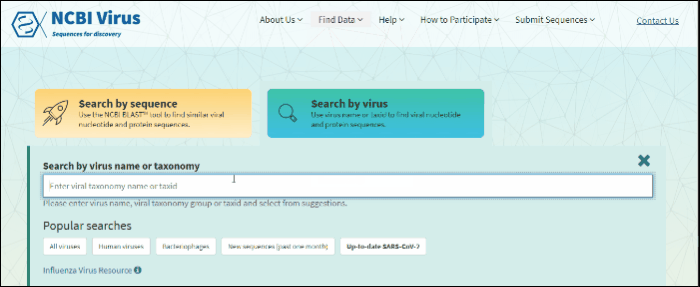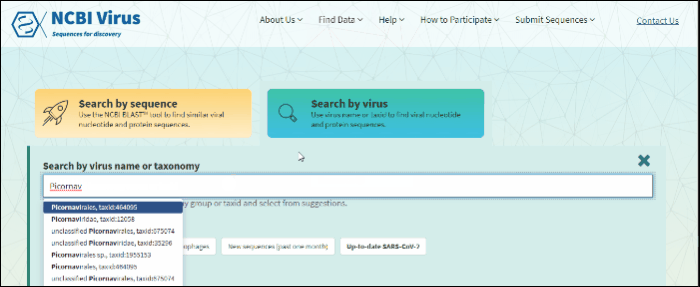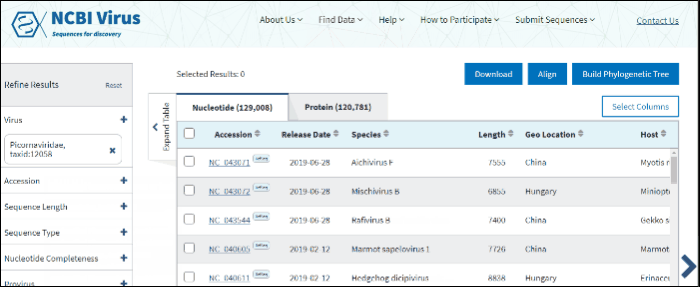
Compare results in the Results Table
Click on the Nucleotide tab to access genomic sequences, the Protein tab to access amino acid sequences for individual proteins, or RefSeq Genome tab to access RefSeq genomes. For segmented viruses each RefSeq genome includes all segments for each segmented virus
In virus search Results Table you can compare search results in tabular display using the following sortable default columns:
-
Accession - the NCBI accession number of the NCBI Virus database sequence.
-
Submitters - authors submitted the sequence. Only the first submitter's name displayed in the column (for example, Baranov,P.V., et al.). To obtain a full list of submitters, click on sequence accession number, this will open the details menu. Click on the accession number in the details panel, this will open GenBank Entrez page with all information available for the selected sequence. Alternatively, you can use the Download button with CSV format option. The column "Submitters" in the downloaded table will contain the name of all authors submitted each sequence.
-
Release date - the date when sequence was released (publicly appeared) in GenBank or other INSDC databases.
-
Isolate - Individual isolate from which the sequence was obtained, typically an alphanumeric sample ID. Isolate name parsed from "/isolate" field of GenBank record. SARS-CoV-2 sequence isolate name is formatted according to the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) definitions.
-
Species – virus species name.
-
Molecule type - viral nucleic acid type. Molecule type is provided by International Committee on Taxonomy of Viruses (ICTV) in the Master Species List and maintained in the NCBI Taxonomy database. RefSeqs that have "Unknown" molecule type belong to tax groups which were not recognized by the ICTV yet.
-
Length - sequence length.
-
Geo Location – country/region of virus specimen collection.
-
USA. If the sample was collected in the United States, the column shows the state abbreviation.
-
Host – virus isolation host (Read more about isolation source vocabulary mapping here). If isolation host is unknown (/host field of the GenBank record), but laboratory host is present (as indicated in /lab_host field of the GenBank record), the laboratory host will be present in the host column of the Results Table. If both isolation host and laboratory host can be mapped, only isolation host will be presented in the host column of the table.
Search results can be customized by adding/removing additional columns from the Results Table in Select Columns dropdown menu.
Additional columns include:
-
Isolation source – sequence isolation source (read more about isolation source here).
-
Collection Date – virus specimen collection date.
-
SRA accession - NCBI Sequence Read Archive (SRA) accession number.
-
Genus.
-
Family.
-
Sequence type – complete/partial/refseq (read more about sequence type here).
-
Nuc completeness - nucleotide completeness (note: it is preliminary data, not always accurate).
-
Genotype.
-
Segment – segment name in case of segmented viruses.
-
Publications - links to associated with sequences publications in PubMed.
-
BioSample – NCBI BioSample accession number.
-
BioProject – NCBI BioProject accession number.
-
GenBank title.
The default number of rows displayed in the Results Table is 200. You can change the number of table rows by selecting number results per page (200, 100, 50 or 25) in Select Columns menu.
Build multiple sequence alignment of selected results
Please, refer to the Build multiple sequence alignment of selected BLAST results, since functionality is the same.
Build phylogenetic tree of selected results
Please, refer to the Build phylogenetic tree of selected BLAST results, since functionality is the same.
Refine tabular results via filters
Please, refer to the Refine tabled BLAST results via filters, since functionality is the same.
How to find, view and download SARS-CoV-2 sequences and related metadata?
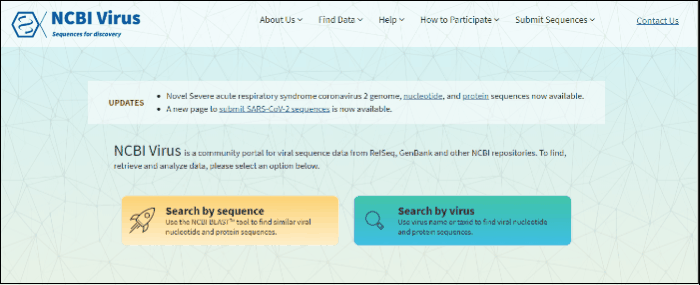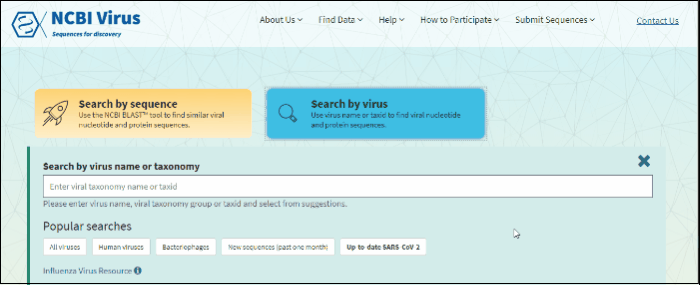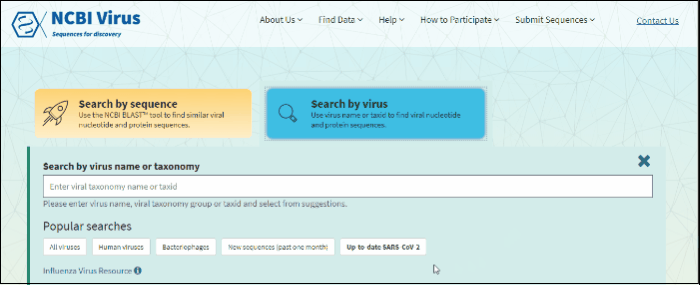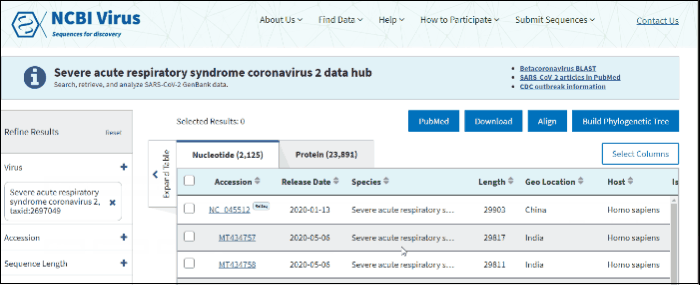
In order to provide free and easy access to genome and protein sequences and associated metadata from the SARS-CoV-2, we created a dedicated Severe acute respiratory syndrome coronavirus 2 data hub.
You can access the Results Table on SARS-CoV-2 data hub, by pressing "RefSeq genomes", "nucleotide" or "protein" links on announcement banner located on NCBI home page, in "Find data" navigation menu or using "Up-to-date SARS-CoV-2" shortcut button in "Search by virus" form.
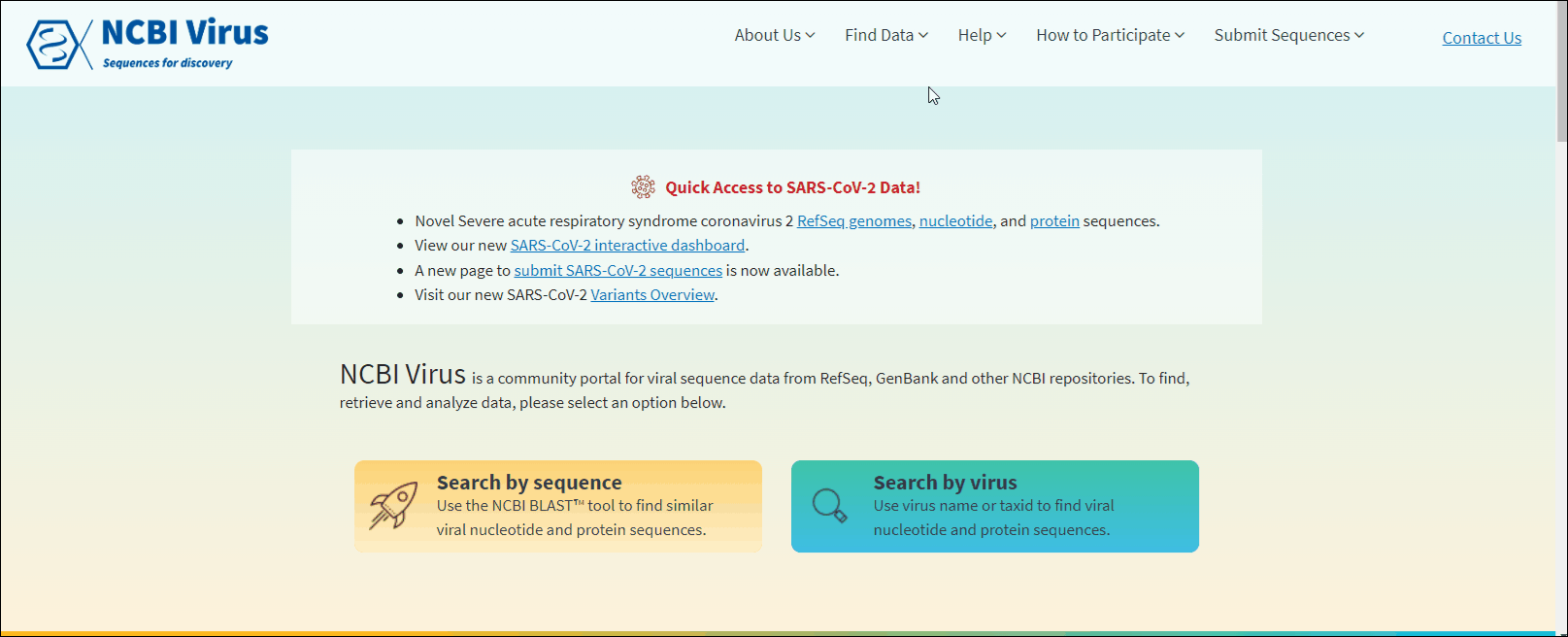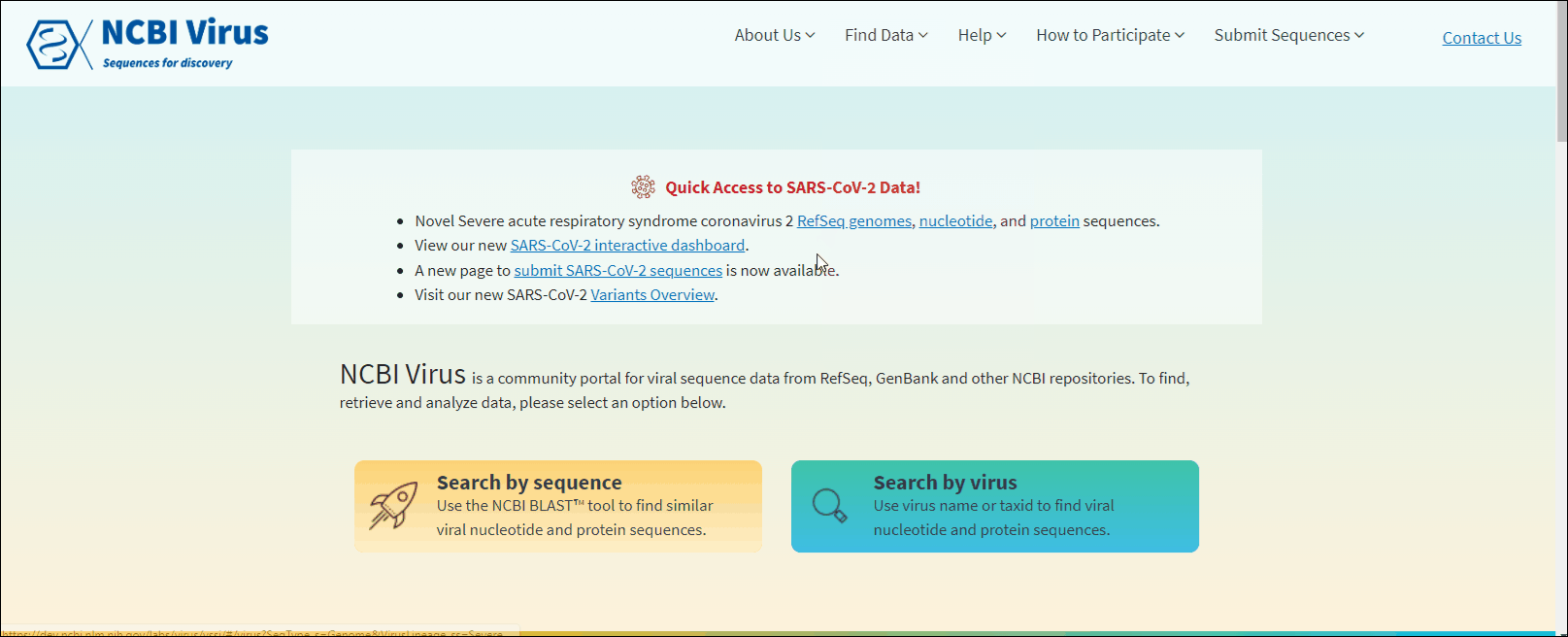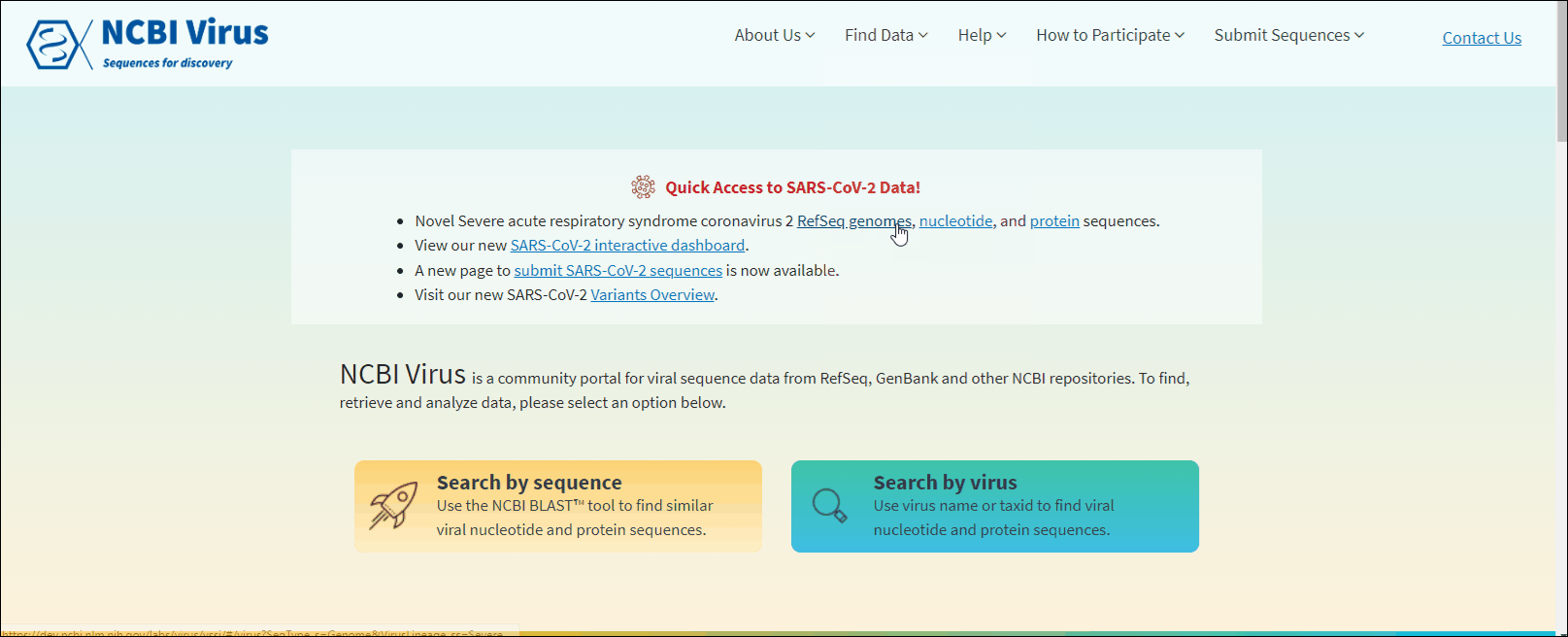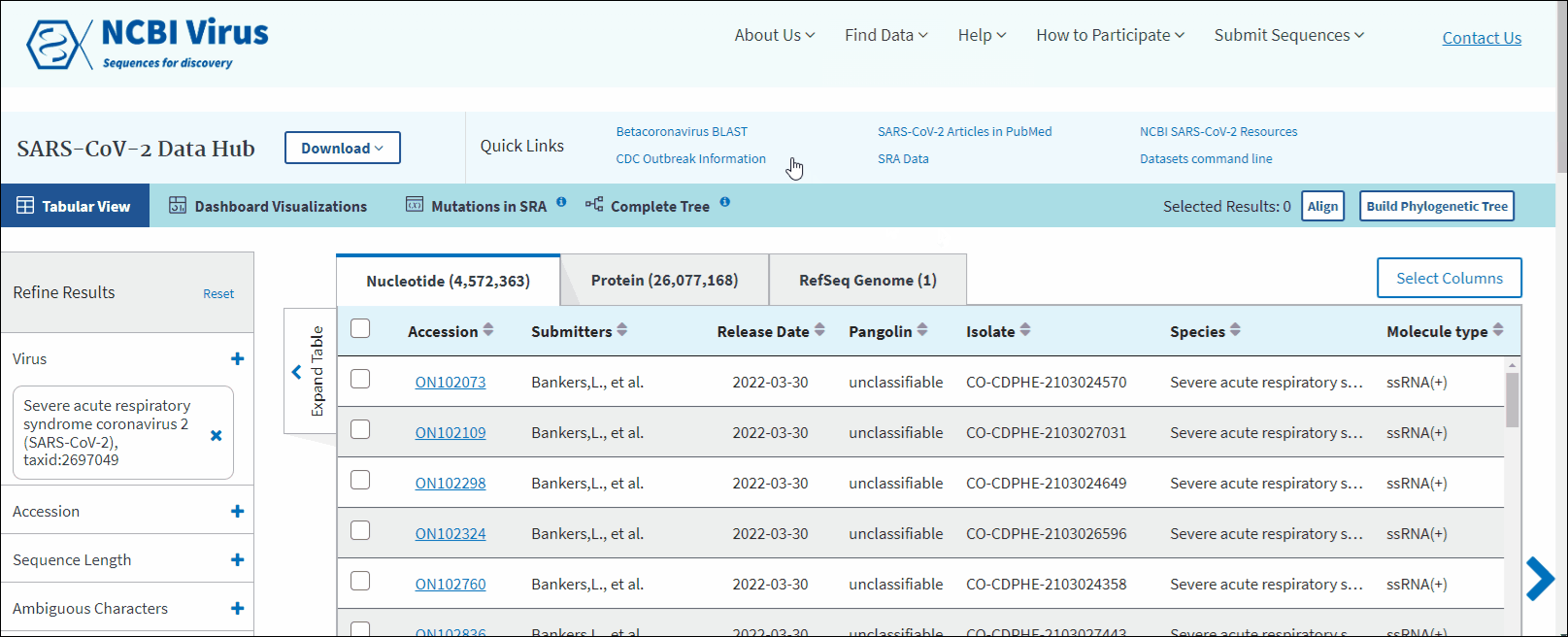
SARS-CoV-2 data hub allows to search, retrieve, and analyze and vizualize SARS-CoV-2 data available in GenBank. This page also provides links to Betacoronavirus BLAST, SARS-CoV-2 articles in PubMed, SRA data, NCBI SARS-CoV-2 resources, Data Sets command line and CDC outbreak information.
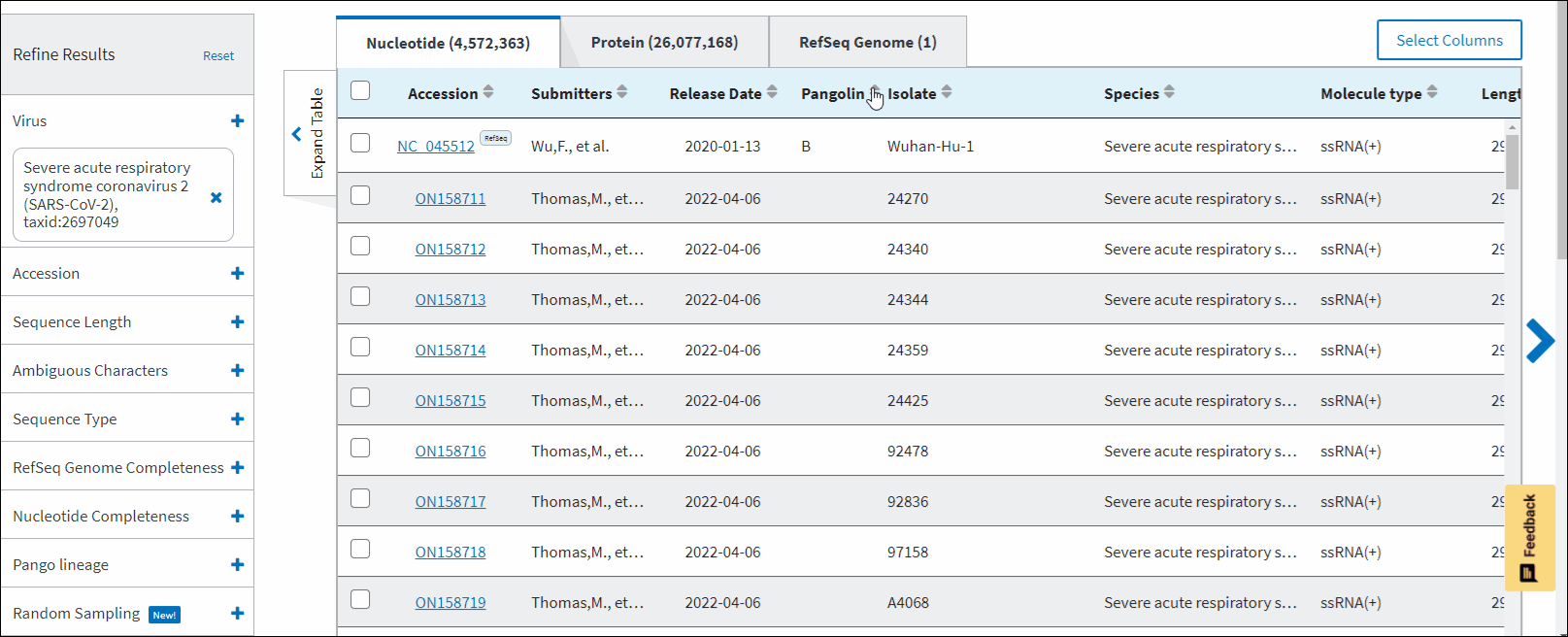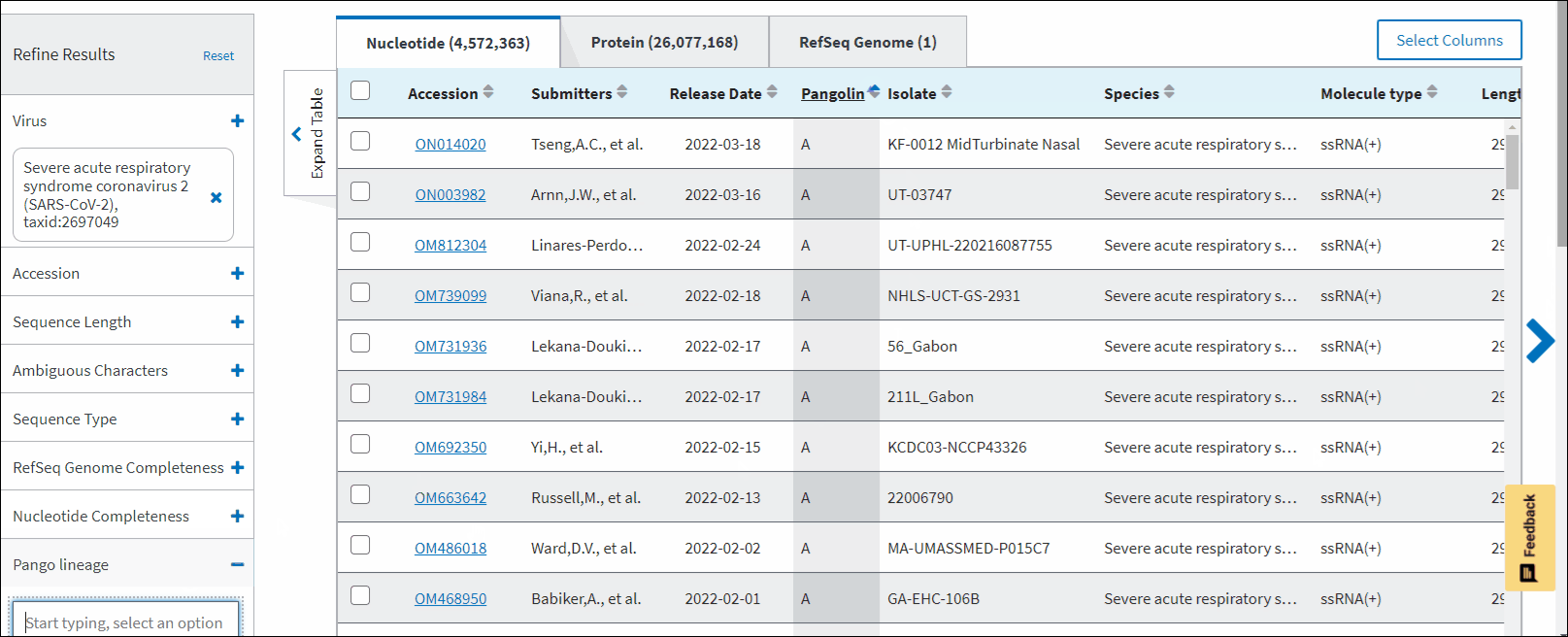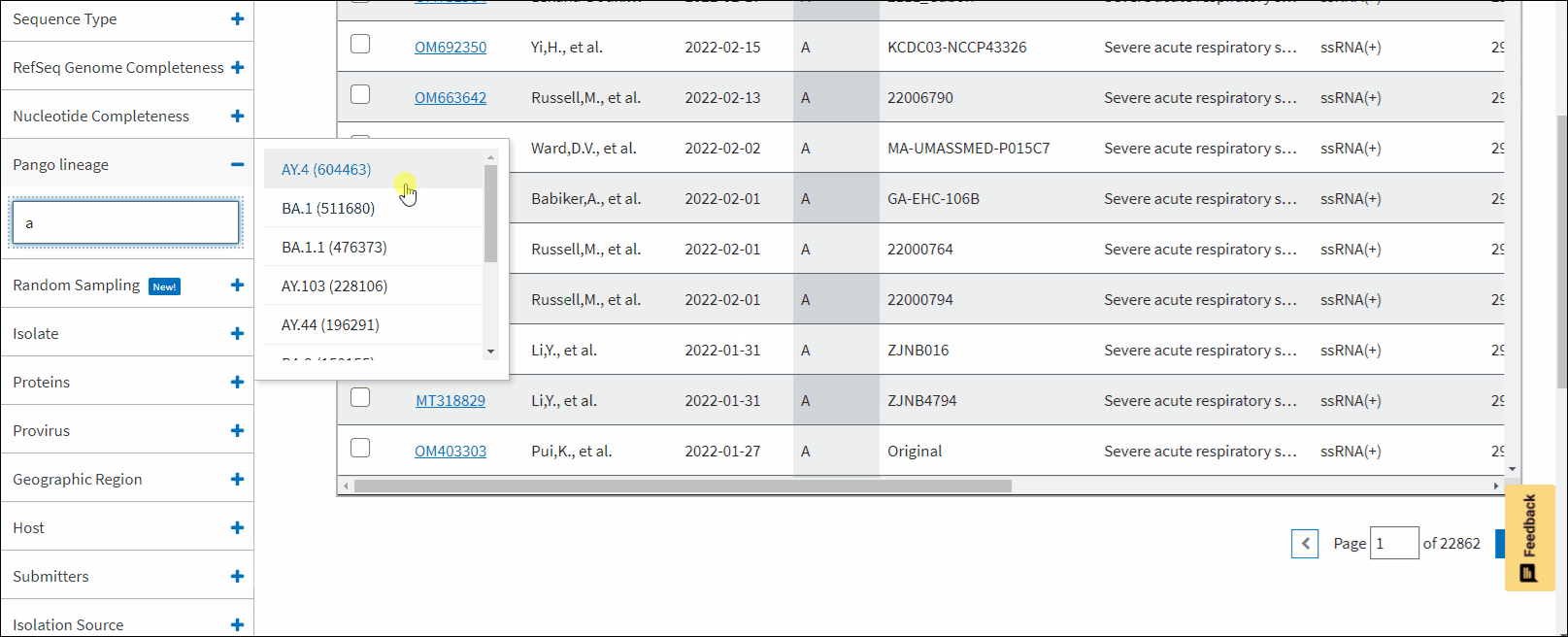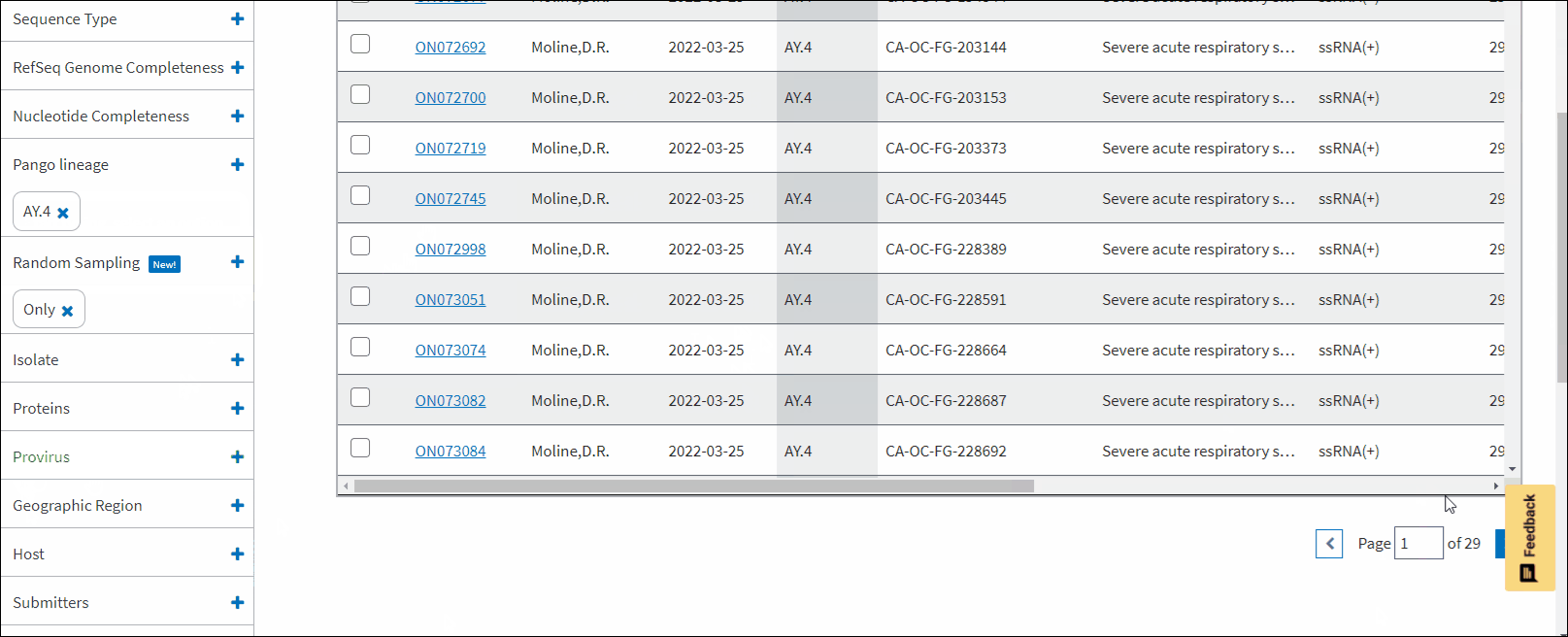
SARS-CoV-2 data hub results table has "Pangolin" column which is specific only to SARS-CoV-2 data. Pango lineages are determined by Pangolin (Phylogenetic Assignment of Named Global Outbreak Lineages). All SARS-CoV-2 GenBank records reprocessed nightly by Pangolin pipeline using UShER pipeline. The field is empty if the sequence was released after the Pangolin run that day. The field will show unclassifiable if the sequence does not meet requirements to be processed, and will show unassigned if the Pangolin tool is not able to determine the lineage for the sequence. You can view Pango version by downloading results in CSV format. You can view version strings in PangoVersions column. Each string includes the following sources: pangolin/pangolin-data/constellations/scorpio. For example, 4.0.6/1.8/v0.1.8/0.3.17.
There are two filters on "Refine Results" panel which are specific only to SARS-CoV-2 data:
-
Pango lineage - allows to filter sequences a particular Pango lineage assigned. Pango lineages are determined by Pangolin (Phylogenetic Assignment of Named Global Outbreak Lineages). All SARS-CoV-2 GenBank records reprocessed nightly by Pangolin pipeline using UShER pipeline. The field is empty if the sequence is unclassifiable or if it was released after a UShER run that day. You can view Pango version by downloading results in CSV format. You can view version strings in PangoVersions column. Each string includes the following sources: pangolin/pangolin-data/constellations/scorpio. For example, 4.0.6/1.8/v0.1.8/0.3.17.
-
Random sampling - allows to filter sequences that were collected randomly for the purpose of baseline surveillance. For example, this filter can be helpful if you would like to know which lineages are increasing in frequency, or are looking for a rough estimate of the infection rate in geographical regions where that data isn’t available yet. Random sampling of samples (e.g., not for vaccine breakthrough or localized outbreak investigation) allows to make these estimates better.
NCBI Virus scanns SARS-CoV-2 GenBank records and any linked BioSample records. If either of the following field/value pairs are found, then the sequence is included in our “random sampling” filter:
- GenBank: KEYWORDS - purposeofsampling:baselinesurveillance
- BioSample: purpose of sequencing - Baseline surveillance
- Influenza virus - allows access to data for the following genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus and Deltainfluenzavirus. Capital letters A, B, C and D in brackets indicate the predominant species in each genus.
- Rotavirus
- Dengue virus
- West Nile virus
- Zika virus
- MERS coronavirus
- Ebolavirus
- SARS-CoV-2 coronavirus
- Please note, our current platform does not have the capability to generate repeatable randomized searches. We realize the importance of repeatability in the scientific community and are working diligently to include this feature in our upcoming updates.
- Downloading randomized subsets in either FASTA format or accession list is currently available for nucleotide, protein, and assembly records. We are working to make them available for coding region records in the future.
- Nucleotide, protein, or coding region sequence (CDS) in FASTA format. Please note, that currently, randomized subsets are not available for coding region sequence (CDS) FASTA files.
- Accession list for nucleotide, protein, or assembly records. Please note, currently, randomized subsets are not available for coding region sequence (CDS) accession lists.
- Results Table – the contents of the Results Table, including the metadata, in CSV format (comma separated values table format) or in XML format.
- only selected records, which were selected using checkboxes in the results table,
- all records in the results table,
- randomized subset of up to 2,000 records in the Results Table (for Nucleotide FASTA, Protein FASTA, Nucleotide Accession List, Protein Accession List, Assembly Accession List, CSV, and XML formats only).
- Assembly
- SRA accession
- Submitters
- Release date
- Pangolin
- Random Sampling
- Isolate
- Species
- Genus
- Family
- Molecule type
- Length
- Sequence type
- Nucleotide Completeness
- Genotype
- Segment
- Publication
- Geo Location
- Country
- Host isolation source
- Collection date
- BioSample
- BioProject
- Dashboard located on the NCBI Virus Home page, which provides virus sequence statistics, Virus Taxonomy Sunburst Chart, and a Host Distribution Bar Chart.
- Dashboard “Visual Filters for GenBank Sequences”, which displays data for specific viral taxa and includes Sequence Type links with calculated virus sequence statistics, a Geographic Distribution choropleth that shows the geographic distribution of sequence records based on collection locations, and time sliders for Collection and Release Date to dynamically show the number of sequences for each time interval.
- RefSeq Nucleotides - all viral nucleotide reference sequences available at NCBI (find more about reference sequences here).
- All Proteins - all NCBI viral protein sequences, including RefSeq proteins.
- All Nucleotides – all viral nucleotide records available at NCBI, including RefSeqs.
- RefSeq Proteins - all viral protein reference sequences available at NCBI.
- Complete Nucleotides – all NCBI viral nucleotide sequences, where GenBank ASN.1 format contains the following descriptors: descr/molinfo/completeness=complete or there is a word 'complete' present in the record’s definition line (defline). It also includes complete reference records (RefSeqs).
- Select ‘Search by Virus’.
- Type virus name, then select an option from the autocomplete list.
- View the results table for your virus of interest.
- Find a tab named “Visual Filters for GenBank Sequences” above the results table.
- Click on the tab “Visual Filters for GenBank Sequences” to switch to visual filtering.
- Click on a selected geographic location to filter sequences by collection location.
- Map allows to select multiple international locations or multiple locations in the USA. The selections will reset if you change between the International and USA maps.
- To select a single location, start typing the name of the region and select the one from a dropdown list.
- Select collection date range of the samples by either selecting one time interval bar or dragging the ends of the sliders.
- Slider displays data from the earliest collection year for this virus data to the current year.
- If the collection time for a record is incomplete, we collapse it like this: If the record only has a year, the record is shown as Jan 1 of that year. If the record only has year and month, the record is shown on the first day of that month.
- Select release date range of the samples by either selecting one bar or dragging the ends of the sliders.
- Slider displays data from the year this virus data was released first time to the current year.
Select Include - to include all randomly sampled SARS-CoV-2 sequences to the Results Table.
Select Exclude - to exclude all randomly sampled SARS-CoV-2 sequences from the Results Table.
Select Only - to view only randomly sampled SARS-CoV-2 sequences.
For other filters description please, refer to the Refine tabled BLAST results via filters, since functionality is the same.
By clicking on "SARS-CoV-2 interactive dashboard" link on the announcement banner located on NCBI home page you can access geographic and time distribution graphs. You also can access it through SARS-CoV-2 data hub
Where can I find SARS-CoV-2 lineage-related information?
You can explore lineage geo-temporal and mutation data using the interactive SARS-CoV-2 Variants Overview dashboard which can be accessed through the announcement banner located on NCBI home page.
Learn more using SARS-CoV-2 Variants Overview help center.
View and download specific virus sequence sets
Find specific data sets
Option 1:
From navigation menu Find data tab select the desired group of viruses: All viruses, Human viruses, Bacteriophages, New sequences (past one month) and Available SARS-CoV-2 sequences to view preselected data sets.
Bacteriophages include virus groups with the following NCBI Taxonomy IDs: 10472, 10656, 10659, 10841, 10860, 10877, 11989, 28883, 1714270, 12333, 79205, 2136181
You can also access the selected virus groups through the "Popular Searchers" panel located on the Results Table. The following virus groups can be accessed:
Option 2:
Click on button Search by sequence located in the central part of NCBI virus home page.
Select the desired popular virus searches group button located beneath the text box.
Both options will open the tabular display with the information about viruses from the selected group.
Learn more how to compare results in tabular display, build multiple sequence alignment of selected results, build phylogenetic tree of selected results or refine the Results Table via filters.
Option 3:
Use NCBI Visual Data Dashboard to explore, view and download the massive, normalized datasets. Learn more.
Download sequences
To download sequences in a variety of formats (FASTA, accession list, the Results Table as CSV or XML), choose Nucleotide, Protein, or RefSeq Genomes tab and optionally select individual sequences to download.
You can also specify if you want to download a randomized or stratified randomized sequence set.
Download a randomized sequence set
Disclaimers
A randomized subset of sequences (also referred to as 'downsampling') can allow a user to work with a smaller subset of sequences selected at random from a larger dataset, as an approximation of the full dataset.
A smaller, representative sequence set could make downstream analysis faster and less computationally intensive, and still allow for interpretation of the larger collection. When downloading a randomized subset, the file name will include the date of download and the randomization seed used.
Examples of file names
| Format | File name |
|---|---|
| FASTA | sequences_[MMDDYYYY]_[seed].fasta |
| Accession list | sequences_[MMDDYYYY]_[seed].acc |
| Results Table - CSV | sequences_[MMDDYYYY]_[seed].csv |
| Results Table - XML | sequences_[MMDDYYYY]_[seed].xml |
Filters can be applied prior to downsampling as described here. After clicking the download button, a menu will allow you to select the download format, then a 2nd step will include an option to download a randomized subset of all the records in your filtered dataset. You can download a set of randomized sequences in a variety of formats (FASTA, accession list, Results table in CSV, or XML formats). Before opening the "Download" menu, please make sure to select the tab above the Results Table which corresponds to the data type you want to download. If you picked the "Nucleotide" tab, you will only be able to download randomized sequence data in FASTA Nucleotide, Nucleotide Accession list, XML, and CSV formats. If you chose the "Protein" tab, you will only be able to download randomized sequence data in FASTA Protein, Protein Accession List, XML, and CSV formats. If you picked the "RefSeq Genomes" tab, you will only be able to download randomized sequence data in Accession Assembly list, XML, and CSV formats.
Download a stratified randomized sequence set
Randomized subsets of sequences can be stratified, meaning equally distributed over a field of categories (also referred to as 'stratified downsampling'). This enables a user to work with a subset of sequences selected from a dataset, as an approximation of the full dataset, with equal numbers of sequences from a selected category, to approximate a larger sequence collection. The fields currently available for stratification are Country and Host. Before opening the "Download" menu, please make sure to select the tab above the Results table which corresponds to the data type you want to download. If you picked the "Nucleotide" tab, you will only be able to download randomized sequence data in FASTA Nucleotide, Nucleotide Accession list, XML, and CSV formats. If you chose the "Protein" tab, you will only be able to download randomized sequence data in FASTA Protein, Protein Accession List, XML, and CSV formats. If you picked the "RefSeq Genomes" tab, you will only be able to download randomized sequence data in Accession Assembly list, XML, and CSV formats.
When downloading a stratified randomized subset, the file name will include the date of download and the randomization seed used.
Examples of file names
| Format | File name |
|---|---|
| FASTA | sequences_[MMDDYYYY]_[seed].fasta |
| Accession list | sequences_[MMDDYYYY]_[seed].acc |
| Results Table - CSV | sequences_[MMDDYYYY]_[seed].csv |
| Results Table - XML | sequences_[MMDDYYYY]_[seed].xml |
Step by step instructions how to download sequences
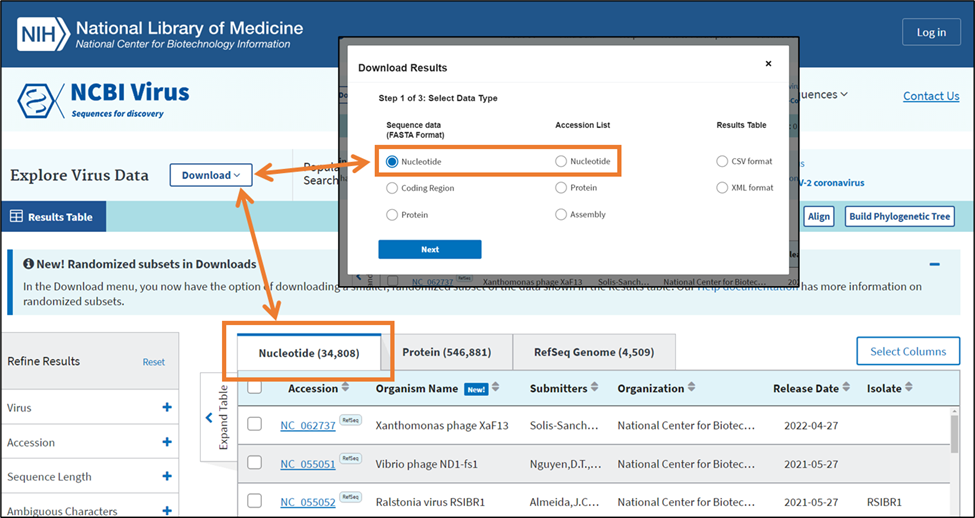
Click Download button on the upper left side of NCBI Virus Results Table page.
This will open the download menu consisting of 3 steps.
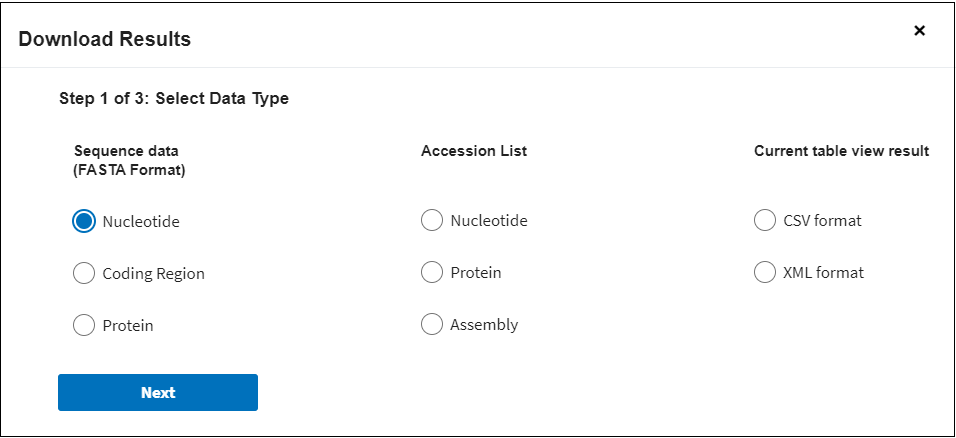
Step 1: Select Data Type.
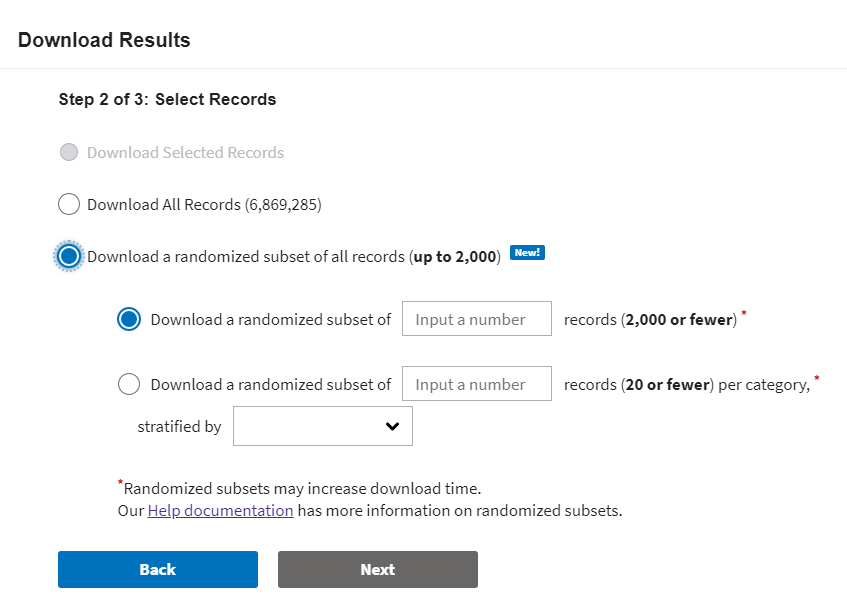
Step 2: Select Records.
Select which records you would like to download:
Randomized subsets contain a limited number of sequences randomly selected from all of the available sequences in the Results Table. As an option, you can choose to stratify your subset by a field, meaning that a roughly equal number of sequences will be randomly selected for each value of that field.
To use options for randomized subsets, select 'Download a randomized subset of all records' and then select either a fully randomized subset or a stratified subset. Enter the total number of randomly sorted records that you want to download into the input box, and enter the category that you want to stratify across from the dropdown.
Randomized subsets contain a limited number of sequences randomly selected from all the available sequences in the Results Table. As an option, you can choose to stratify your subset by a field (up to 20 records country or per host), meaning that a roughly equal number of sequences will be randomly selected for each value of that field.
To use options for randomized subsets, select 'Download a randomized subset of records (up to 2,000) and then select either a fully randomized subset or a stratified subset. Enter the number of randomly sorted records (up to 2,000 for randomized subset and up 20 records per value for stratified subset) that you want to download into the input box and enter the category that you want to stratify across from the dropdown.
The fields currently available for stratification are Country and Host.
Click “Next” and follow the prompts on the 3rd step in the menu to begin your download.
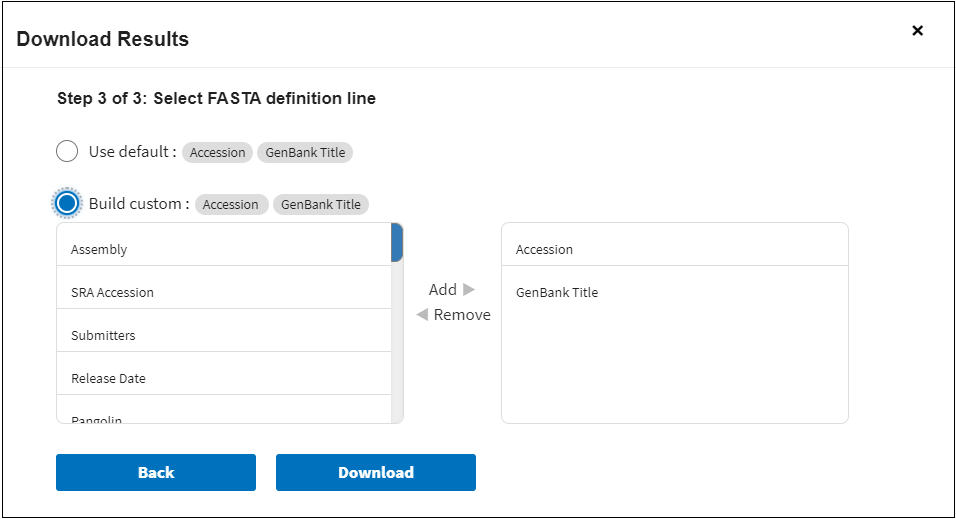
Step 3.
If in step 1 you selected Sequence Data (FASTA format), in step 3 you can select FASTA definition line for the sequences that you are going to download.
In case if nucleotide or protein sequence data were selected in Step 1, the default FASTA definition line will be presented in the format (accession) | (GenBank title) and will include the GenBank sequence accession number and GenBank title:
>AAO17794 |VP4 spike protein[Human rotavirus A].
In case if coding region option was selected, the default definition line format will be (nucleotide accession)_(cds coordinates)_| (GenBank title) and will include the related GenBank nucleotide sequence accession number, the indication that this is a coding region (cds), related GenBank protein accession number and related protein GenBank title:
>NC_045425.1:319..1659 |replication endonuclease [Thermus phage phiOH3].
You can change this default defline to fit your own needs by selecting Build custom sequence title option. Here you can select the following options (columns):
You can view description for each option in the description of the Results Table columns.
If in Step 1 you selected the Accession list , you can download nucleotide, protein and and RefSeq genome assembly accession numbers with or without vesrsion number. For example: NC_045512 (without version) or NC_045512.2 (with version).
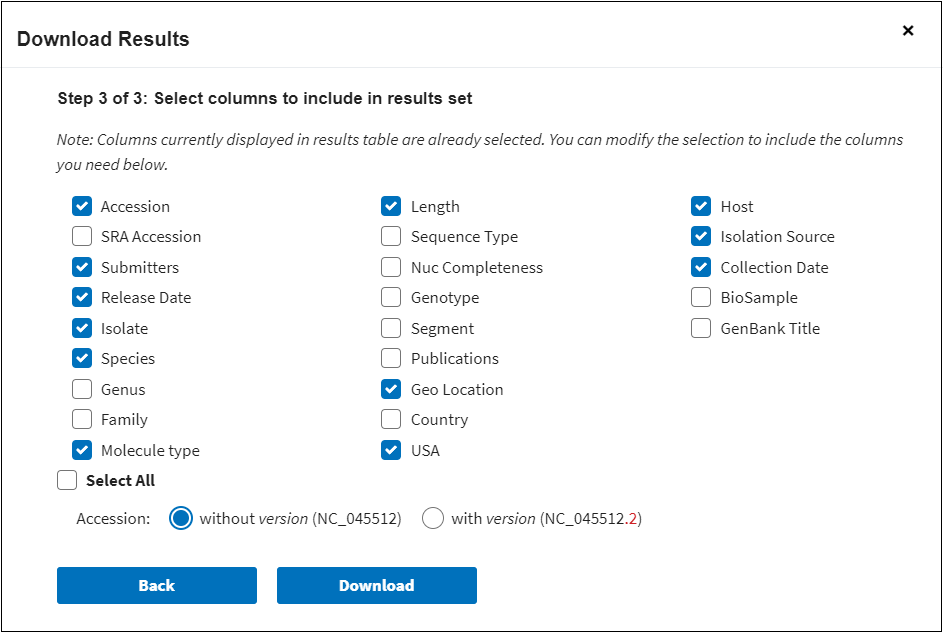
If in Step 1 you selected the the Results Table in CSV format, the downloaded results will show all selected columns data. You can modify the selected columns and choose the columns you need in Step 3: Select columns to include in results set. You also can select if you want to include accession number with or without version number.
NCBI Visual Data Dashboards
NCBI Virus visual data dashboards support data exploration and discovery across our normalized datasets. They can be used to identify trends in data and to select specific subsets based on those trends.
Visual dashboards in Virus encompass:
See the instructions below for how to use each dashboard.
1: Home Page Dashboard
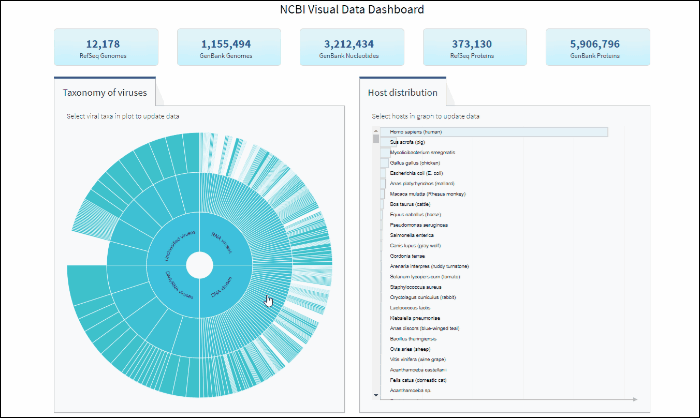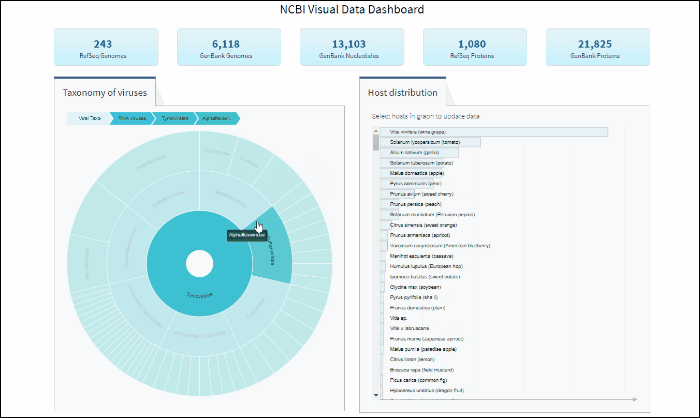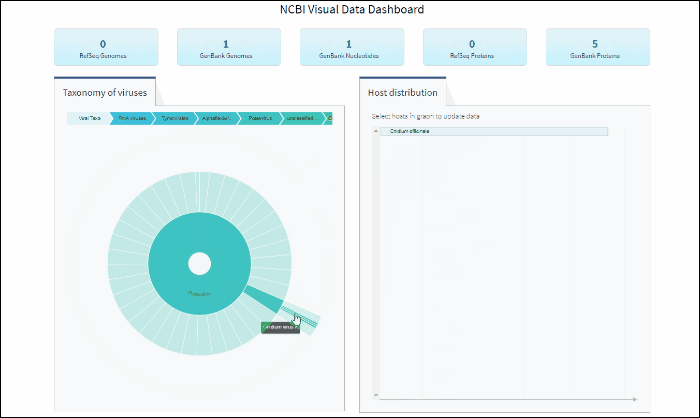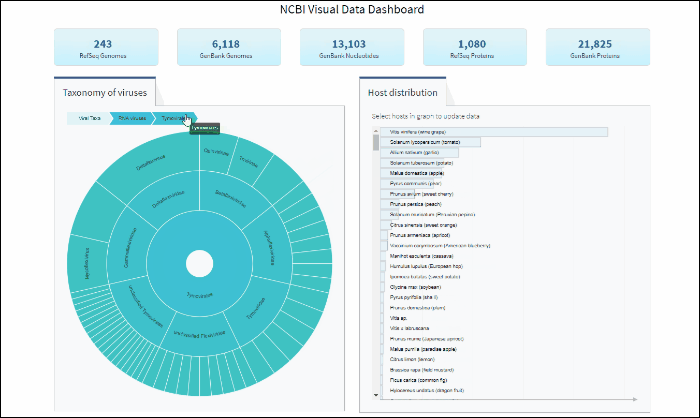
Access sequence data via buttons located in the top row for the following statistics:
Clicking on each button will show a results table with the corresponding sequences. Those results can be further refined by using filters for various sequence attributes (metadata) located on the left side of the Results Table page (learn more here).
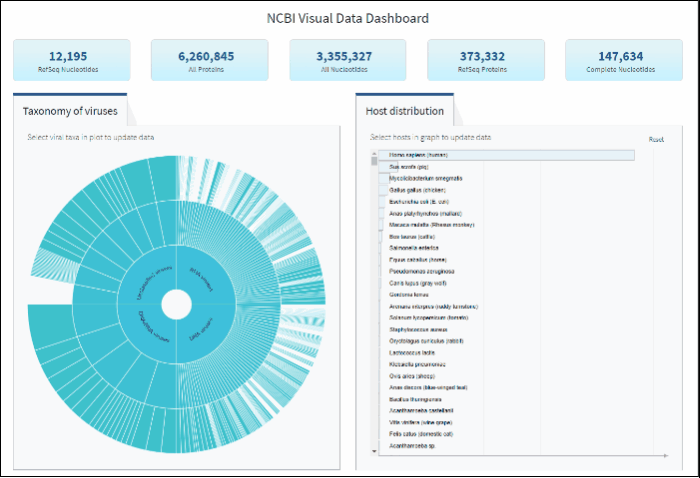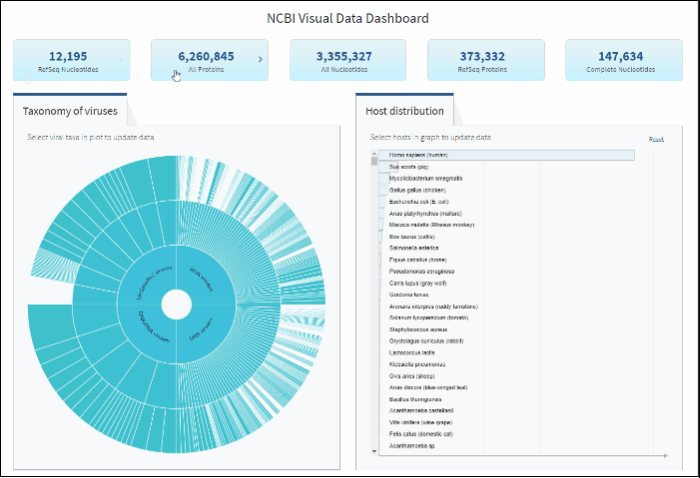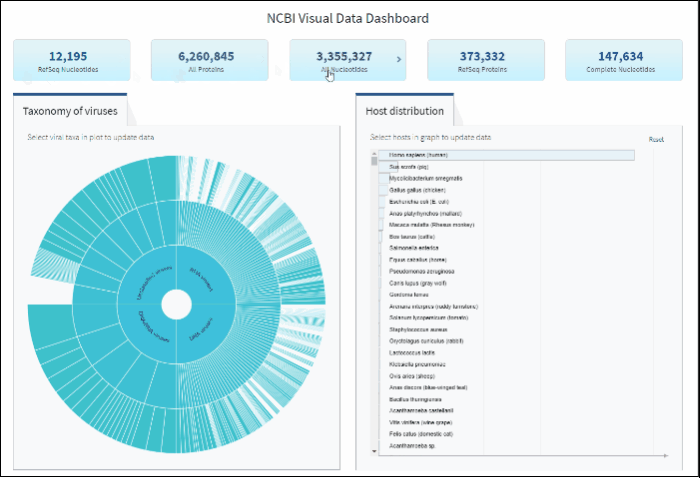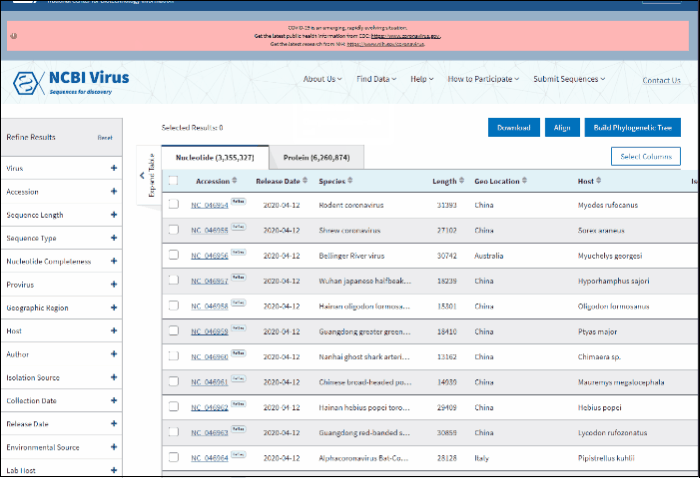
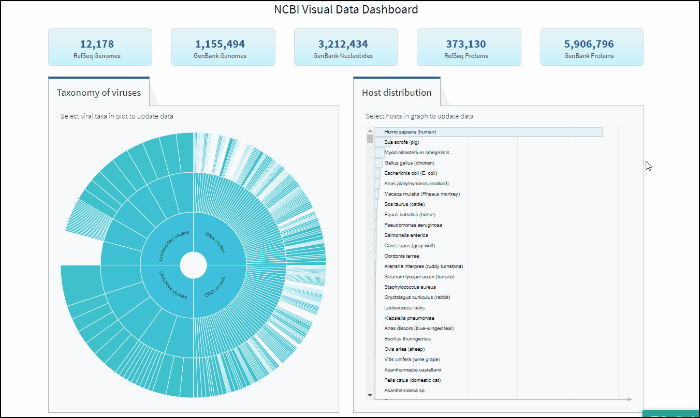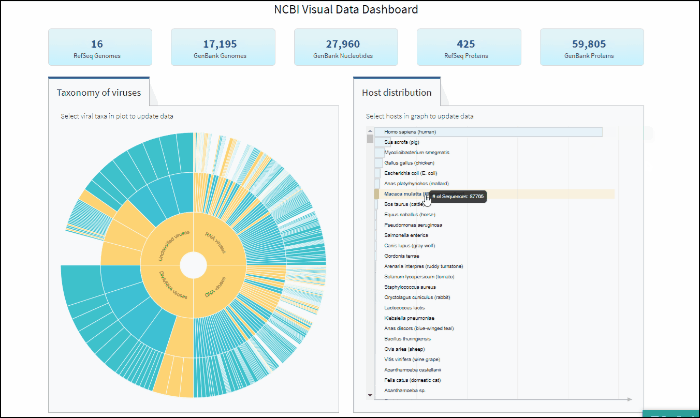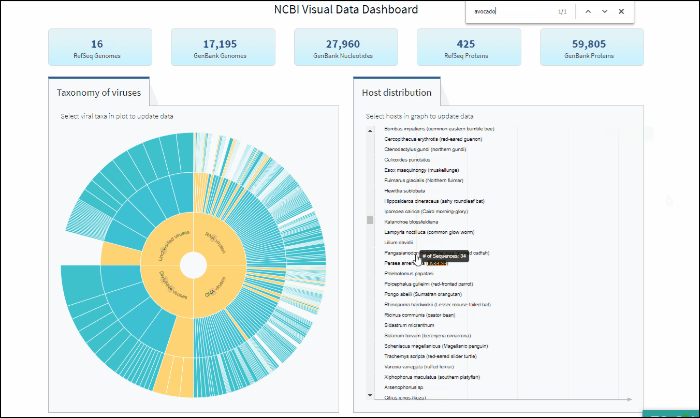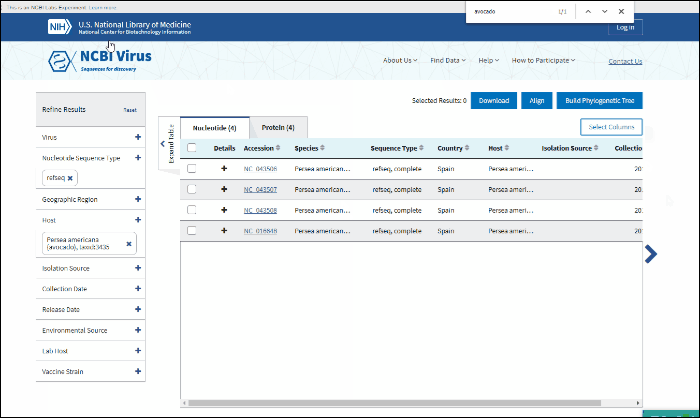
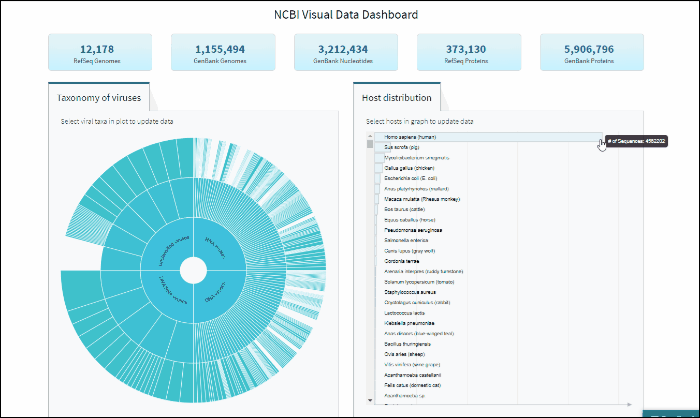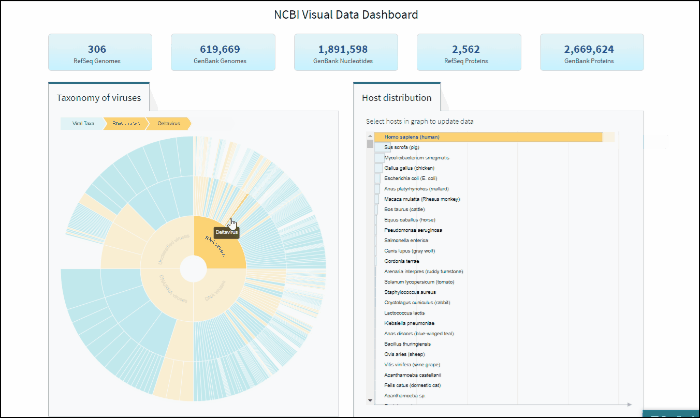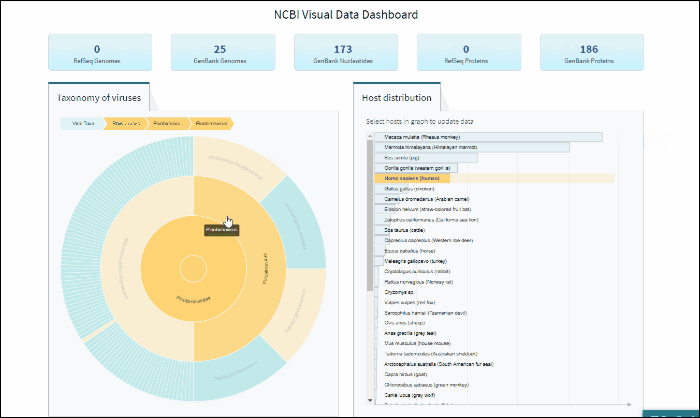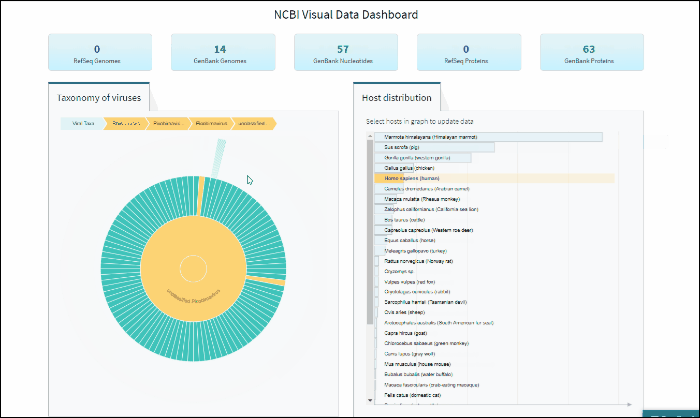
Explore virus taxonomy hierarchy using sunburst chart
Virus taxonomy can be explored via an interactive sunburst chart. The default view represents the classification for all available NCBI viral taxa. The inner layer (ring) represents four non-taxonomic groups of viruses: RNA viruses, DNA viruses, DNA/RNA viruses (which includes reverse-transcribing viruses), and Unclassified viruses. Only 4 levels of the whole hierarchy are visible on the plot at a given time.
To explore virus taxonomy, click on any slice (section) of any layer on the sunburst chart. This will trigger the plot to zoom into the selected taxa and display any additional taxa below the selection. Each viral taxa name is displayed on a corresponding slice or can be viewed in the hover-over tool-tip by placing your cursor over the slice. Dynamic breadcrumbs with viral taxa names are located above the sunburst plot. Breadcrumbs are also a secondary navigation system that show the location of the taxa in the hierarchy and clicking on one will refocus the plot on the selected taxa. You can also see breadcrumbs by hovering over any slice in the sunburst. Clicking on the center of the sunburst chart will return you to the parent taxa.
Select specific virus taxonomy group and view statistics for specific sequence sets with quick links to download them
After selecting a specific taxonomy group on sunburst chart, you can view and explore the updated statistics in the top row of the dashboard.
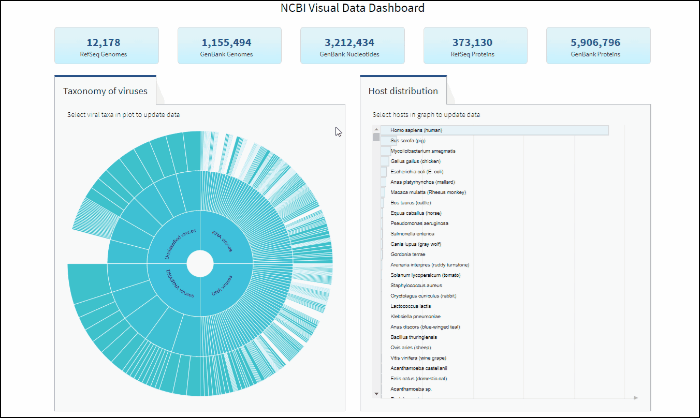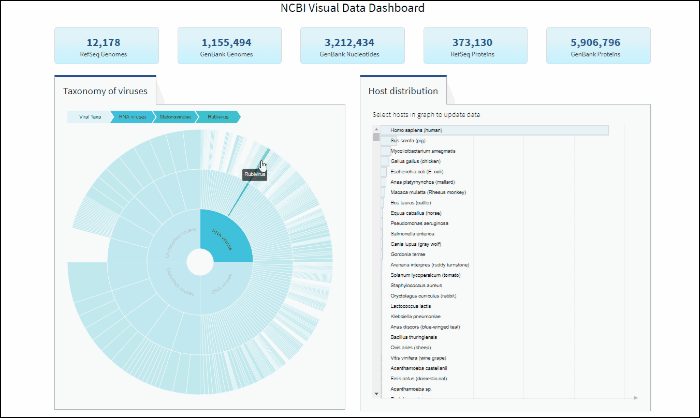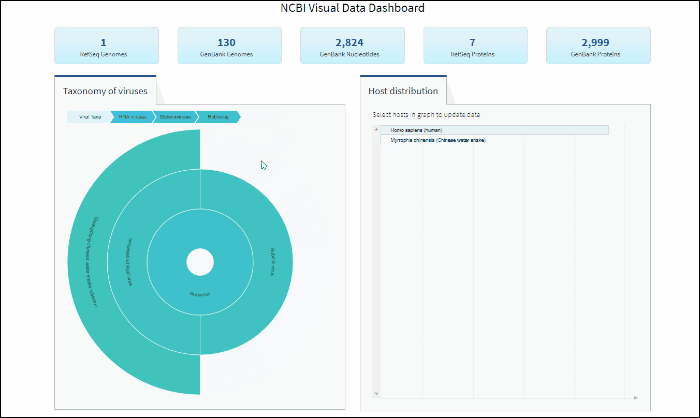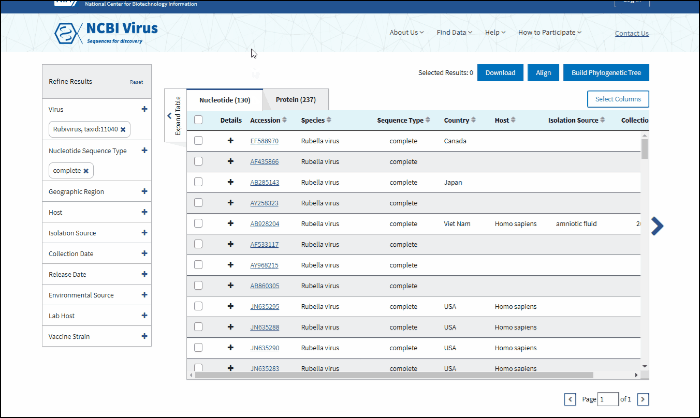
Select a host term from the Host Distribution bar chart and see the distribution of that host among the various viral taxa
The interactive Host Distribution chart shows the distribution of virus host species. Each host bar is proportional to the number of virus sequences isolated from this host. The total number of virus sequences for each bar can be viewed by hovering over the bar.
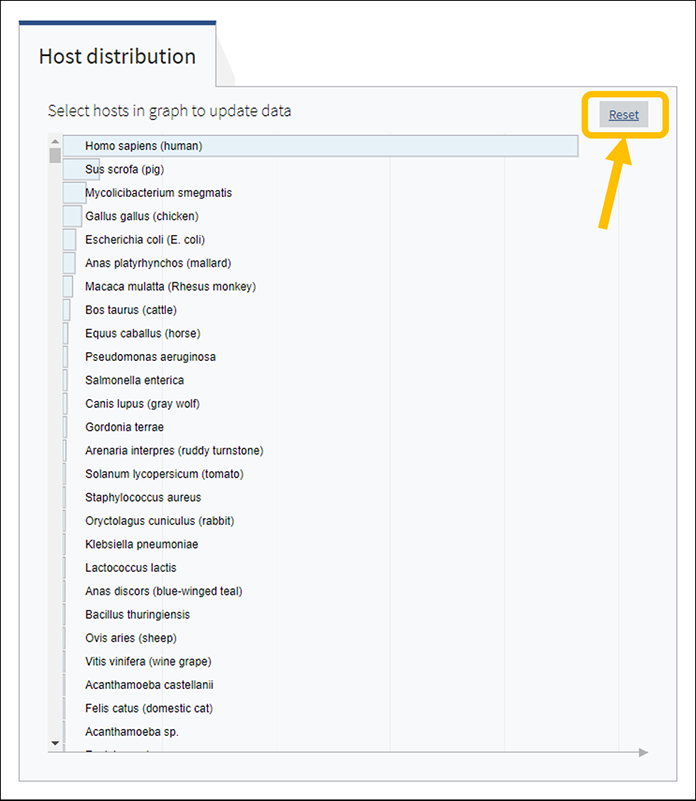
To select a host species, click on a bar or on a corresponding host name. This will highlight selected host, as well as all virus taxonomy groups containing sequences isolated from the selected host. Only one host can be selected at a time. Clicking on the selected host a second time will de-select it or you can use the Reset option available in the top right corner of the host chart. The statistics in the top row of the dashboard will be updated based on the selected host.
You can search for a host species by scrolling the scrollbar on Host Distribution Chart, or by using keyboard combination "CTRL+F".
You can reset Host Distribution chart the the original view by pressing on button "Reset" in the upper right corner of the chart.
Explore viral taxonomy hierarchy within a given taxon highlighted by the host selection
By clicking on a highlighted taxonomy group, you can further explore viral taxonomy hierarchy on sunburst chart. The lower layers that include taxa with sequences from the selected host will be highlighted. While zooming in, not all taxa will be highlighted if not all taxa include sequences from the selected host.
2: “Visual Filters for GenBank Sequences” Dashboard
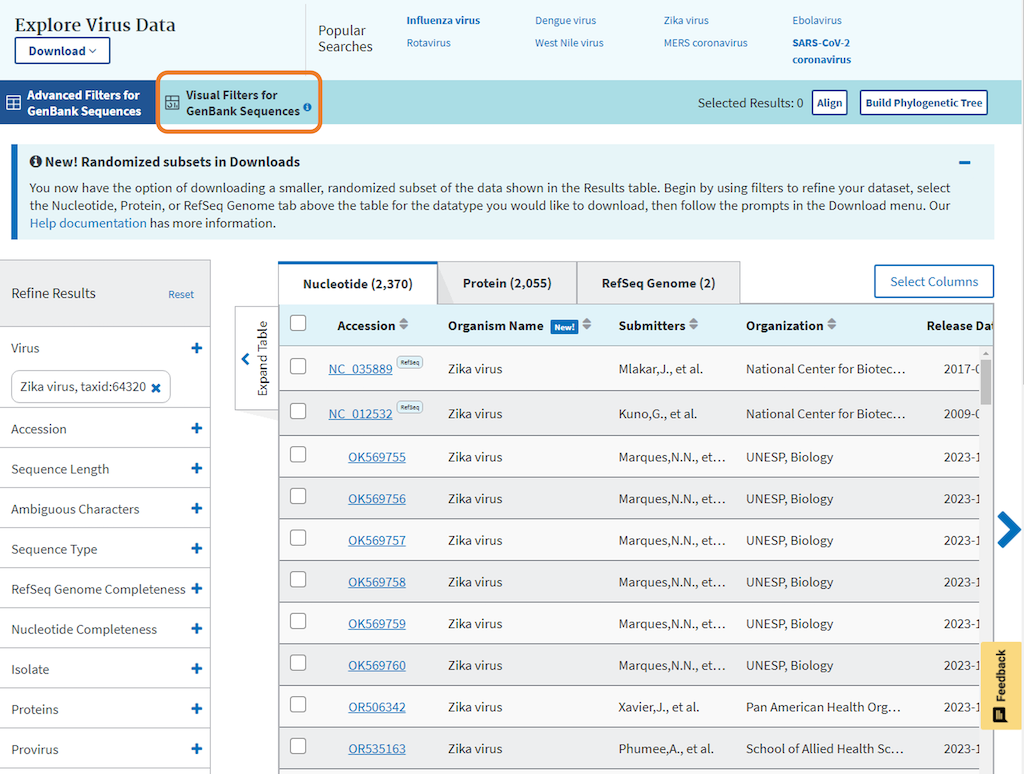
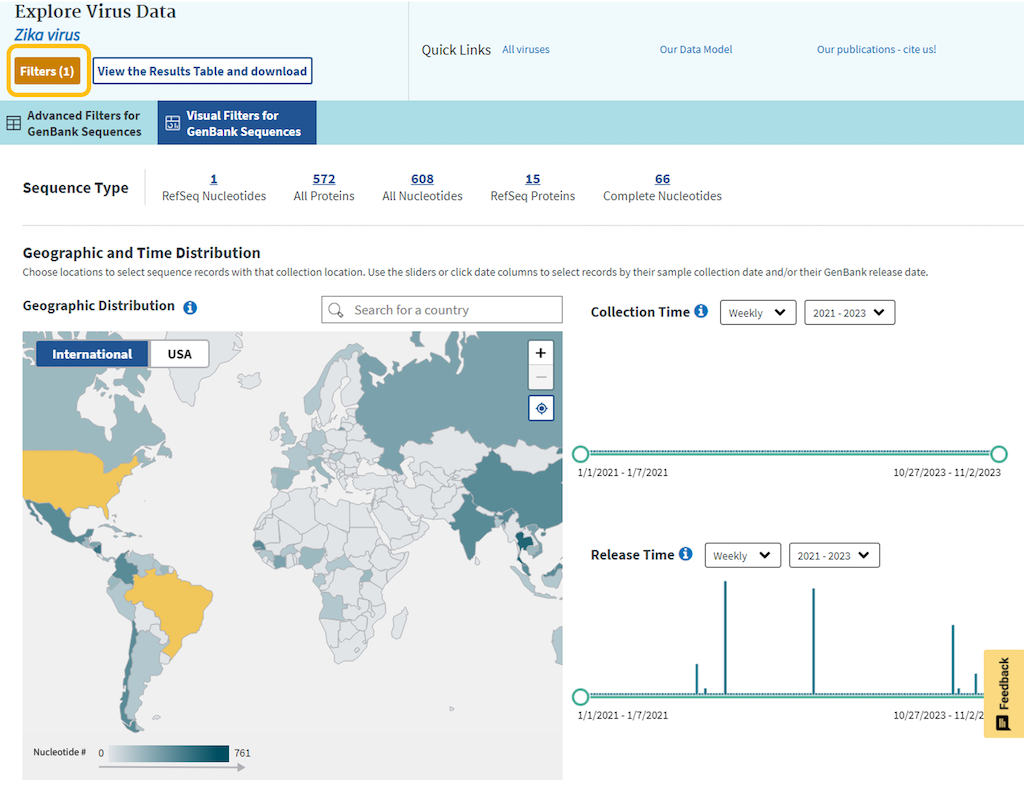
“Visual Filters for GenBank Sequences” is a dashboard which enables filtering of your virus search results based on important attributes, like geographic location, collection, and release date, using visualized, graphical filters.
How to access “Visual Filters for GenBank Sequences”?
There are several ways to access Visual Filters for GenBank Sequences.
1. From NCBI Virus home page follow the steps below:
2. From the Results Table page access the “Visual Filters for GenBank Sequences” tab in the header above the results table.
Please note, if any filters were applied on the results table, switching to the “Visual Filters for GenBank Sequences” dashboard will reset all the filters except for the virus name.
3. By adding NCBI Virus “taxid” number directly to the page URL: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/dashboard?taxid=
For example, for Zika virus (taxid=64320), enter the following URL: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/dashboard?taxid=64320
How to use Visual Filters for GenBank Sequences?
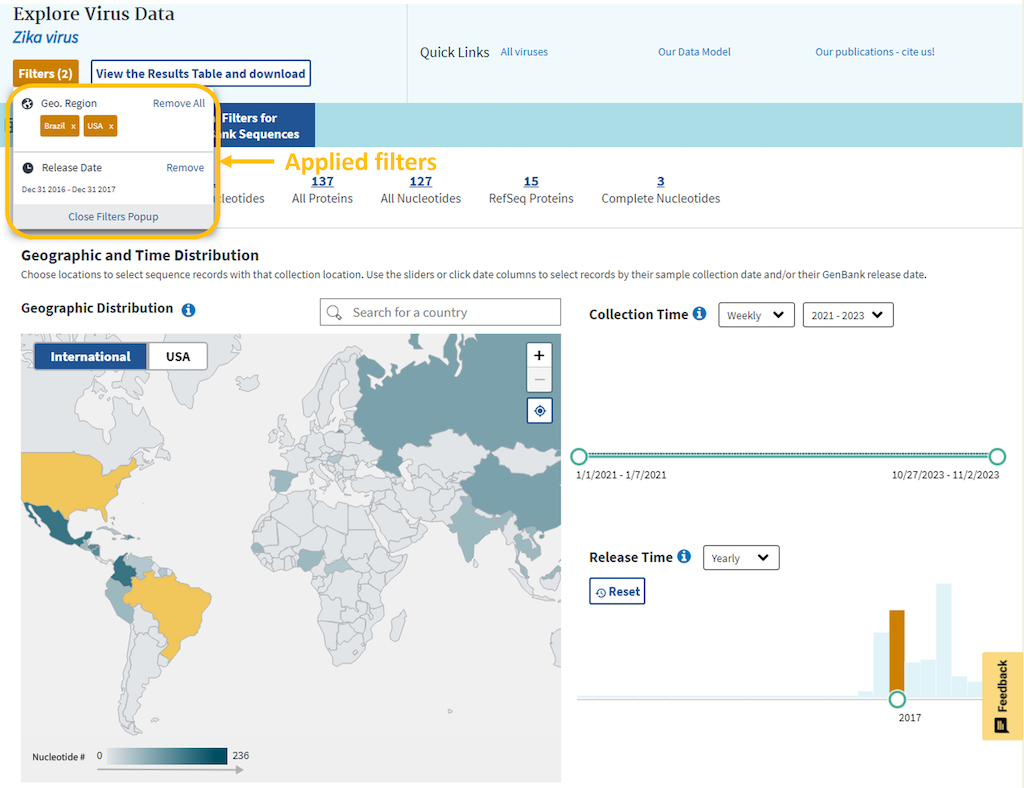
Visual filters allow to filter your search by geographic location, collection time, and release time. Each filtering feature on the dashboard is interactive and connective, so when a filter is applied in one feature, it is also reflected in the other features. When using these filters, the top summary section is automatically updated to reflect the number of records in the NCBI RefSeq, Nucleotide, and Protein sets in the NCBI Virus database that fit the combined conditions of your search.
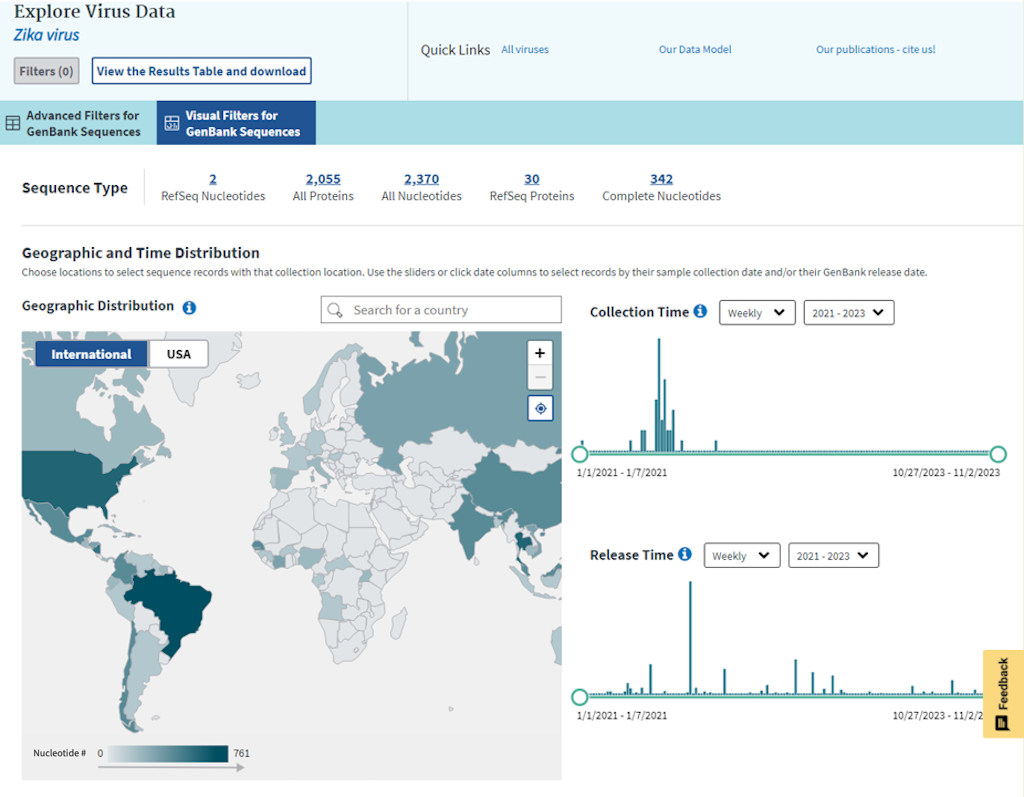
Geographic Distribution choropleth map allows to select sequence records collected at that location.
Please note, that color shades on the map are based on nucleotide record numbers for the virus; darker shades correspond to higher numbers, and lighter shades - to lower numbers.
By using the Collection Time and Release Time sliders, you can view a histogram of distribution of nucleotide record numbers in different time intervals.
Use the sliders or click date columns to select records by the sample collection date or the GenBank release date. Weekly, monthly and yearly time intervals can be selected.
Collection Time graph:
Release Time graph:
You can also select different bi-yearly intervals, which will show you the portion of the graph for that time frame. However, you still have to click on the bar or select the time interval with the sliders to apply filtering.
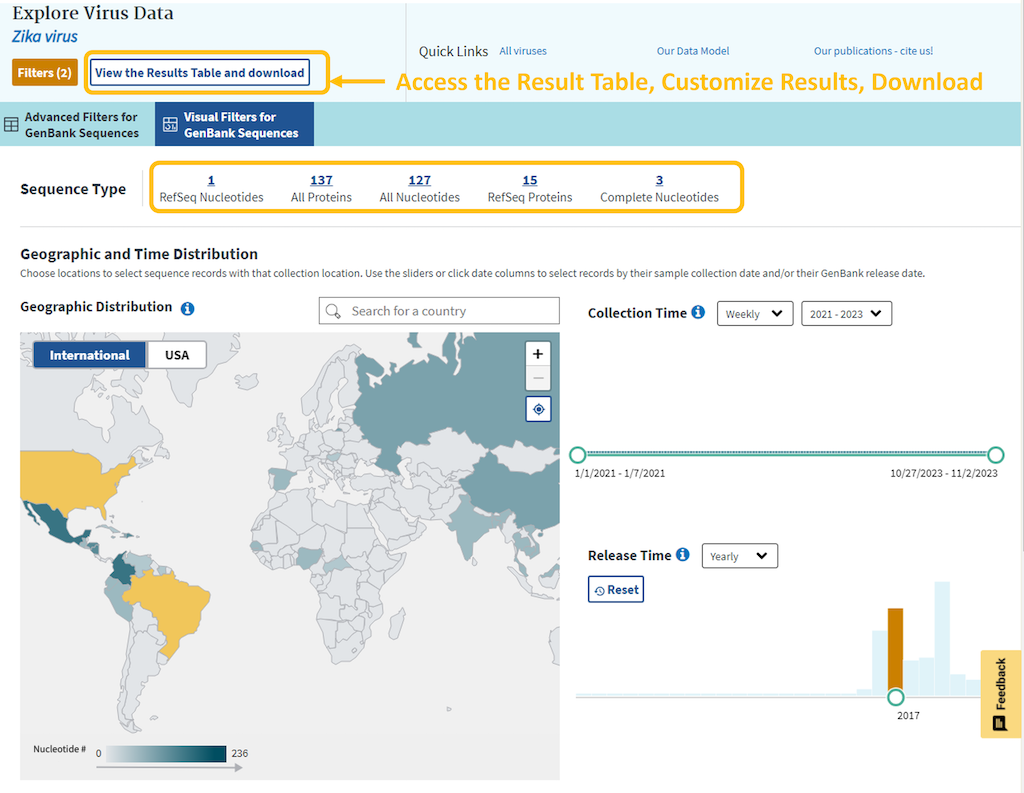
The top header of the Dashboard includes a link back to the Results Table page where you can review your results in tabular format, apply more filters, and download FASTA sequences, an accession list, or the table itself.
Note, that all filters applied in the graphical view will remain in effect on the Result Table page. However, if you switch from the Results Table page back to the visual filters, all applied filters will be lost, except for the selected virus name.
How to find, view and download HIV-1 sequences and related metadata?
Public HIV-1 nucleotide and protein sequence data are displayed in HIV-1 data hub.
HIV-1 data hub can be accessed by typing and selecting HIV-1 in Search by virus name or taxonomy input form.
Alternatively, it can be accessed from NCBI home page by typing HIV-1 in search window. This will open another page with HIV-1 virus genome assembly information. Press on NCBI virus button to access HIV-1 data hub.
These are early days for HIV-1 data support in NCBI Virus. Please stay tuned for updates and further details relevant to HIV-1.