NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
SNP FAQ Archive [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005-.
This publication is provided for historical reference only and the information may be out of date.
Searching for dbSNP Statistics
Can you show me the growth rate of dbSNP for all organisms through the present (end of August 2007)?
Can you show me the overall growth of human variations in dbSNP over the years since its inception?
How do I determine the number of validated indel SNPs in dbSNP human b127?
How do I find the number of rsSNPs that map uniquely to the human genome?
How do I determine the number of SNPs there is per gene across all genes in the human genome?
Searching for Data Histories and Historical Data
Could you tell me the number of validated and unvalidated SNPs in dbSNP build 120?
How do I locate information for two fairly old submitted SNPs: ss35527513 and ss35527511?
Can you show me the overall growth of dbSNP over the years since its inception?
Finding a Build Number Based on a Date and Vice Versa
Where do I find the dates when a particular SNP was released for public view?
Searching for SNP Entry Dates for Patent Purposes
Searching for dbSNP Statistics
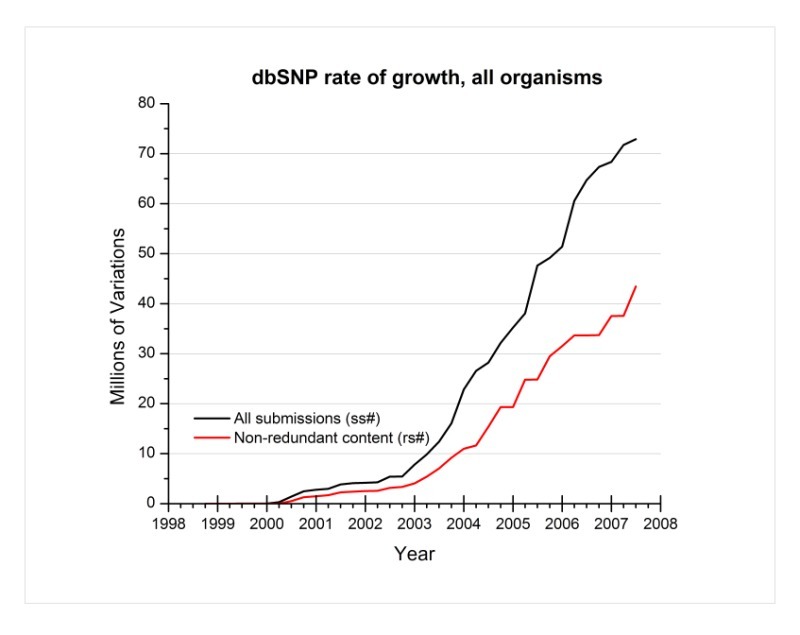
Can you show me the growth rate of dbSNP for all organisms through the present (end of August 2007)?
Below is a graph of dbSNP’s growth rate for all organisms. The black line indicates the growth rate of dbSNP using the total number of submissions, while the red line indicates the growth rate using the non-redundant content (refSNPs) of dbSNP.
(08/28/07)
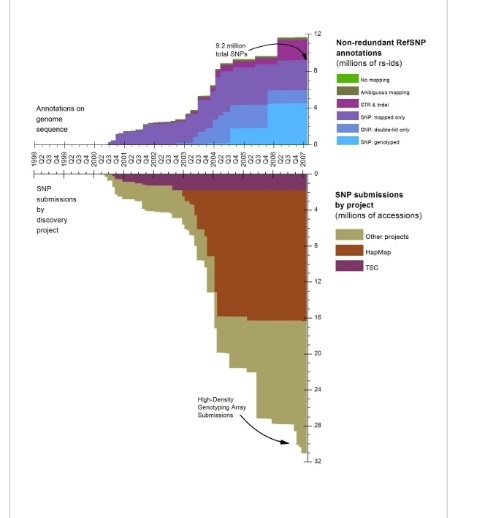
Can you show me the overall growth of human variations in dbSNP over the years since its inception?
Below is a graph of the growth of dbSNP human through February, 2007. At that point we were closing on 12 million non redundant variations clustered over 30 million submissions.
(2/27/07)
I would like to cite the number of SNPs reported today in the human genome. Can you point me to a good reference?
Go to the dbSNP summary page, and scroll down to the BUILD STATISTICS table. (7/6/07)
What proportion of SNPs reported in dbSNP has been identified through the international HapMap project?
As of this date, dbSNP has individual genotyping data from HapMap project for 4.1 million SNPs out of the 10+ million total SNPs in dbSNP. dbSNP currently has genotype data from HapMap release 21.a. (10/9/07)
How do I determine the number of validated indel SNPs in dbSNP human b127?
- 1.
Go to EntrezSNP.
- 2.
Select the “Limits” tab located just below the textbox at the top of the page.
- 3.
Go to the limits section called “SNP class”, and select “in del".
- 4.
Go to the limits section called “Validation Status”, and select the following:
"by-frequency"
"by submitter"
"by 2hit-2allele"The default response will show the results for all organisims in dbSNP. To see the results for human or mouse, click on the grey tabs just below the yellow bar that displays the limits you selected.
Using this search, 12245 human refSNPs that meet the criteria you selected as of today.
A table describing the validation codes is available online. (3/27/07)
What percentage of SNPs deposited in dbSNP is located in coding regions (exons) versus non-coding regions (introns + intergenic regions)?
You can get these numbers by querying Entrez SNP:
- 1.
Type "All[sb]" (without the quotes) in the search text box located at the top of the page.
- 2.
Click on the “Preview/Index” tab located just below the search text box.
- 3.
Scroll to the bottom of the Preview/Index page, and click the drop-down menu toggle arrow just to the right of “All Fields”. Select "Function Class" from the list.
- 4.
Click on the "Index" box.
- 5.
A list of SNP functional classes will appear with the number of SNPs in each functional class located to the right of each functional class. (6/21/05)
How do I find the number of non-synonymous SNPs in dbSNP?
To get the number of non-synonymous SNPs, click on the geneReport link located next to HOMO SAPIENS under Build Statistics on the build summary page.
How do I find the number of rsSNPs that map uniquely to the human genome?
Conduct the query on Entrez SNP:
- 1.
Enter the following query in the Search box to search for human SNPs that map once to the genome and not on unknown chromosomes:
- 2.
(human AND 1[WEIGHT]) NOT un[CHR]
- 3.
Change the query weight to 2 for the SNPs that map twice.
How do I determine the number of SNPs there is per gene across all genes in the human genome?
Use the ContigLocusId table, which contains all the SNPs mapped to genes in the human genome. The ContigLocusID table is located in the organism data directory of the human database in the bSNP FTP site. (9/2/05)
Searching for Data Histories and Historical Data
Broad’s Haploview shows HapMap genotype data along with SNP data mapped to assembly 35. Current dbSNP data is mapped to build 127 (assembly 36). How do I use dbSNP to map a list of refSNPs to assembly 35?
dbSNP does not support searches for older assembly coordinates. You can get coordinates from dbSNP build 125 and 126, which were based on genome assembly 35 from the SNPContigLoc tables for those builds. You will find these tables in the organism_data subdirectory, which is located in the human database directory of dbSNP’s FTP site.(9/28/07)
Could you tell me the number of validated and unvalidated SNPs in dbSNP build 120?
Build120 was released on Mar 16 2004 12:00AM and mapped to the NCBI Human Genome 34 ver. 3. Build 120 has a total of 9,098,790 human SNPs, and 4,267,639 of those are validated.
Please note that dbSNP validation status is delineated by various levels. You can see definitions for the various validation status levels by clicking on the words "Validation Status" located on any RefSNP page in the table header just below the banner that reads “Submitter records for this RefSNP Cluster”. (3/14/05)
Where can I find the history of dbSNP mouse submissions — specifically those from Celera, as well as other commercial and non-commercial labs?
- 1.
Go to the dbSNP home page.
- 2.
Locate the Submission Information section and select the “New Batches” link.
- 3.
Select “mouse_10090” from the organism dropdown menu and then choose Select “Variation submissions (SNP)” from the Batch Type dropdown menu
- 4.
Click the Search Batch button at the bottom of the page, and you will be given a list of SNP batch submissions that includes submitter handle, batch ID, and the number of variations submitted in descending calendar order. (2/21/06)
How do I query dbSNP so that it will return a flat or xml file containing the new RefSNP (rs) ID number into which a previously valid rs recently merged?
You can get the rs merge history of all rs numbers from your organism’s (human in this case) RsMergeArch table located in on the dbSNP ftp site.
The following example shows that rs4344934 has been merged to rs1107123:
gzcat RsMergeArch.bcp.gz | grep 4344934
4344934 1107123 123 1 2004-09-24 18:49:00 2004-10-10
11:55:00 1107123 1Also, you can retrieve a list of merged rs numbers from Entrez SNP. Just type “mergedrs” (without the quotation marks) in the text box at the top of the page and click the “go” button. Each entry in the returned list will include the old rs numbers that has merged, and the new rs number it has merged into (with a link to the refSNPpage for the new rs number). You can limit the output to merged rs numbers within a certain species by clicking on the “Limits” tab and then selecting the organism you wish from the organism selection box. (5/25/05:11/03/08)
Some RefSNP (rs) numbers in dbSNP are merged into one rs number. Where does dbSNP provide the merge history?
You can now retrieve a list of merged rs numbers from Entrez SNP. Just type “mergedrs” (without the quotation marks) in the text box at the top of the page and click the “go” button. Each entry in the returned list will include the old rs numbers that has merged, and the new rs number it has merged into (with a link to the refSNPpage for the new rs number). You can limit the output to merged rs numbers within a certain species by clicking on the “Limits” tab and then selecting the organism you wish from the organism selection box.
You can also use the RsMergeArch table located in the dbSNP FTP site. (11/8/05:11/03/08)
How do I locate information for two fairly old submitted SNPs: ss35527513 and ss35527511?
Search for these ss numbers by using the “Search by ID” query module located on the dbSNP homepage:
- 1.
Enter the ss ID number in the input text box.
- 2.
Select "NCBI Assay IDs" from the drop down menu to the right of the input text box.
- 3.
Click “Search”
- 4.
You will get a “Submitted SNP(ss) Report” for the ss ID you entered for your search.
- 5.
Click on the blue ss Id number located at the top of the report to go to the submitted SNP page for this ss ID number, or click on the blue rs Id number also located at the top of the report to get refSNP mapping information. (10/14/05)
Can you show me the overall growth of dbSNP over the years since its inception?
Below is a graph of the growth of dbSNP human through February, 2007. At that point we were closing on 12 million non redundant variations clustered over 30 million submissions.
(2/27/07)
Finding a Build Number Based on a Date and Vice Versa
I downloaded dbSNP on Jun 6, 2004, and now need to provide a reference for this build in a manuscript. Can you tell me the build number?
It looks like it was dbSNP build 121. If you scroll to the bottom of the SNP summary page, you can query there for release dates from the most recent build all the way back to build 106. (1/19/05)
Where do I find the dates when a particular SNP was released for public view?
You can find these dates by checking the build history page for the release dates (these immediately follow the submission dates. PLEASE NOTE that the dates found in the NCBI databases are not to be used for patent purposes. See the NCBI Copyright and Disclaimer page for additional information.
Searching for SNP Entry Dates for Patent Purposes
Our Patent Office asked us to supply the Issue Date, Entry Date, and any updates for two SNPs in dbSNP. How do I get this information?
The dates found in the NCBI databases are NOT to be used for patent purposes. See the NCBI Copyright and Disclaimer page for additional information.
You can find dates when a particular SNP was released for public view by checking the build history page for the release dates (these immediately follow the submission dates. (10/14/08)
- Searching for Statistics, Data Histories and Historical Data in dbSNP - SNP FAQ ...Searching for Statistics, Data Histories and Historical Data in dbSNP - SNP FAQ Archive
Your browsing activity is empty.
Activity recording is turned off.
See more...