NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
SNP FAQ Archive [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005-.
This publication is provided for historical reference only and the information may be out of date.
Statistics on Total Number of dbSNP Variations
Can you show me the growth rate of dbSNP for all organisms through the present (end of August 2007)?
How do I find out the number of SNPs submitted to dbSNP so far?
Why is the total number of refSNP (rs) IDs less than the total number of submitted SNP (ss) IDs?
Number Validated Statistic
Number of SNPs Identified by HapMap
Number of SNPs Mined from Literature
What percentage of SNPs in dbSNP has been mined from the literature?
Number of Submitted SNPs with Frequency Statistic
Number of indel SNPs
Why did the total number of indels in the human genome go down between dbSNP b126 and b127?
Number of Triallelic SNPs
Calculations of Mean SNP Density
Where do I find recent calculations of SNP mean density in the human genome?
Total number of RefSNPs vs.Total Number of Submitted SNPs
Summary of Annotated dbSNP Entries
dbSNP Growth Rate Statistics
Can you show me the overall growth of dbSNP over the years since its inception?
Statistics on Total Number of dbSNP Variations
According to dbSNP, is it safe to say that as of June, 2008, there are 12.8 million SNPs or single base genetic variations in the human genome to date?
Yes. In build 128, dbSNP has about 12 million uniquely mapped refSNP (rs) numbers. Please note that dbSNP contains not only has single base variations, but also contains indesl, STRs, and MNPs(multiple base nucleotide variations). To see all the variation classes available in dbSNP, you can go to the EntrezSNP limits page (click on the “limits” tab located just below the search boxes at the top of the page).
In the current build, 129, the number of uniquely mapped refSNP (rs) numbers has grown to about 14 million+. (06/10/08)
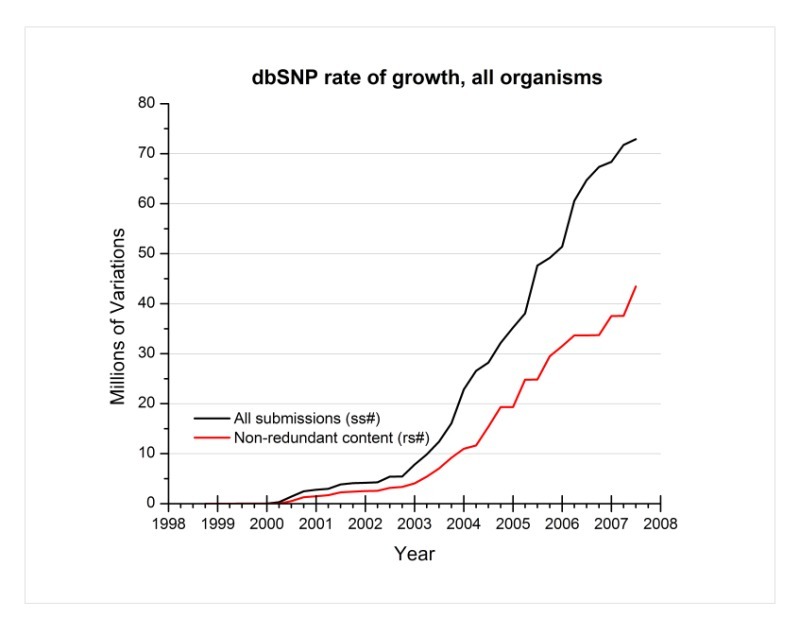
Can you show me the growth rate of dbSNP for all organisms through the present (end of August 2007)?
Below is a graph of dbSNP’s growth rate for all organisms. The black line indicates the growth rate of dbSNP using the total number of submissions, while the red line indicates the growth rate using the non-redundant content (refSNPs) of dbSNP.
(08/28/07)
How do I find out the number of SNPs submitted to dbSNP so far?
You can get a summary for each organism online. (8/11/05)
I would like to cite the number of SNPs reported today in the human genome. Can you point me to a good reference?
Go to the dbSNP summary page, and scroll down to the BUILD STATISTICS table. (7/6/07)
What is the current number of SNPs in the human genome?
As of September, 2006 there are 9,669,384 true SNPs (I exclude microsatellites and MNPs from our database in the search) in our database that map exactly once to a well-defined location on the NCBI build 33 human genome.
You can determine this yourself. Start here.
- 1.
Check all chromosomes except W and unknown in the Chromosome(s) box.
- 2.
Check 1 in the Map weight box (to get just the unique mappings).
- 3.
Check Homo sapiens in the Organism(s) box.
- 4.
Check SNP in the SNP class box.
- 5.
Now, go to the top of page and enter “human[orgn]” (without the quotation marks) in the Search box, then click on the Preview/Index tab located directly underneath the Search box, and once you are in the Preview/Index section, click the Preview button. SNP counts will be displayed as active links that will take you to a list of available SNPs.
This list will be too long to be useful, so you may want to go back to the Limits page and refine your search.(9/13/06)
Does dbSNP have a table that shows all the variations ever submitted to dbSNP along with their frequencies? If this data is available, how do we separate it into synonymous and non-synonymous changes?
The various base pair changes submitted to dbSNP are located in a table called UniVariation.
There currently is no database table that stores the frequencies of the different variations, but various queries can be constructed that will find this information.
For example, the query result below shows the count of human SNPs with each variation pattern for a single nucleotide substitution. The total SNP count in human is 10430639. This query is based only upon the variations themselves; it does not address the frequency that those variations occur in (a) population(s). Are you making a distinction between G>T and T>G in your question?
_
univar_id variation rs_cnt 2 A/G 3178537 5 A/C 879718 23 A/T 747669 25 G/T 887321 35 C/T 3174225 40 C/G 862671 Below is a query result that reports variation counts separated by synonymous and non-synonymous changes.
_
var_st snpFunction rs_cnt A/C synon 2573 A/C nonsynon 5923 A/G synon 18613 A/G nonsynon 19031 A/T synon 1051 A/T nonsynon 3226 C/G synon 3077 C/G nonsynon 7950 C/T synon 20440 C/T nonsynon 17786 G/T synon 2297 G/T nonsynon 5869 The total human synonymous SNP count is: 48261
The total human non-synonymous count is: 60725
(2/2/06)
Why is the total number of refSNP (rs) IDs less than the total number of submitted SNP (ss) IDs?
We group multiple submitted SNPs (ss) that are the same into a cluster and assign a unique, non-redundant reference SNP (rs) number to that cluster of submitted SNPs. (3/10/06)
Number Validated Statistic
What is the definition of the b125 and b126 build statistic for the “# validated” figure? Which validation methods does this statistic include?
You can find the definition for this term on the dbSNP glossary page.. (7/13/06)
Number of SNPs Identified by HapMap
What proportion of SNPs reported in dbSNP has been identified through the international HapMap project?
As of this date, dbSNP has individual genotyping data from HapMap project for 4.1 million SNPs out of the 10+ million total SNPs in dbSNP. dbSNP currently has genotype data from HapMap release 21.a. (10/9/07)
Number of SNPs Mined from Literature
What percentage of SNPs in dbSNP has been mined from the literature?
As of this date, only a very small percentage (< %1) of the variations in dbSNP are mined from literature, but starting later this spring, we expect a greater percentage of SNPs will come from the literature as dbSNP will have two new resources that will allow users to submit previously published human variations described in a publication. These resources are the Human Variation: Annotate and Submit Batch Data site (for multiple submissions) and the Human Variation: Search, Annotate, Submit site (for single submissions). The submission forms for these resources allow you to link the variation you are submitting to the PubMed ID number of the original publication, so the original reference for the variation discovery is maintained. (05/15/08)
Number of Submitted SNPs with Frequency Statistic
What is the definition of the b125 and b126 build statistic for the number of Submitted SNPs (ss#s) with Frequency?
You can find the definition for this term on the dbSNP glossary page. (7/13/06)
Number of indel SNPs
Why did the total number of indels in the human genome go down between dbSNP b126 and b127?
There are 2202926 indel SNPs in b126, while there are 2145084 indel SNPs in b127. The reason why the b127 indel SNP count went down is that many of these SNPs ( a total of 943011) were found to map to the same genome locations after the release of b126. These SNPs were merged during the mapping process for b127. (3/27/07)
Number of Triallelic SNPs
Can you tell me what proportion of SNPs in dbSNP are triallelic, and what proportion of these are triallelic because of copy number variation?
Please note that out of the total 14-15 million SNPs in the human database, we have a total of 5133 STR SNPs with various repeat numbers. (e.g. a SNP has variation as: (CA)7/8/9/10/15/17)
I am not a biologist, and so when I did this query, I assumed a variation such as "(A)14/15/16" could be called a microsatellite, an STR(short tandem repeat), or a CNV(copy number variation).(05/12/08)
Number of SNPs per Gene
Has anyone used dbSNP to determine the median number of nonsynonymous SNPs per gene and the most nonsynonymous SNPs in a gene?
There have been a number of studies that examined nonsynonymous SNPs in the human genome.
Try searching PubMed using “nonsynonymous”, “SNP” and “genome” or other search terms as key words. (08/28/08)
Calculations of Mean SNP Density
Where do I find recent calculations of SNP mean density in the human genome?
Go to the dbSNP summary page. Then click on geneReport for the organism of interest in the organism statistics table. If you click on geneReport for human, you will be connected to the human SNP density report.
Total number of RefSNPs vs.Total Number of Submitted SNPs
Shouldn't the number of new refSNPs be smaller than the number of new submitted SNPs? If so, why are there so many new human rs clusters in B126?
A large number of submitted SNPs did not get clustered in the build 125 release, and were assigned refSNP numbers for them in build 126. There were 2.3 million newly assigned refSNP numbers in build 126, most of which (600,000) are just new clusters of existing submitted SNPs that were submitted in build125.
In the ideal dbSNP world, all SNPs submitted during the time span of a build would be clustered for the release of that build. But in the real dbSNP world, we load submitted SNPs on a daily basis, and by the time all the submitted SNPs are mapped, clustered, and the frequency information for them computed, we end up with many new submitted SNPs that have not been assigned refSNP numbers. We are working to improve the build pipeline, and hope to reduce the time lag between new submitted SNPs loading, and refSNP number assignment.(5/24/06)
Summary of Annotated dbSNP Entries
Where can I find a summary of annotated human dbSNP entries (i.e. total number of SNPs, indels, cd-indels, 5'UTR indels, 3' UTR indels… etc.).
Sorry we don't have this information in a summary, but you can use Entrez SNP to query for the counts:
- 1.
Go to Entrez SNP
- 2.
Select the grey “Limits” tab, located near the top of the page just below the search text boxes.
- 3.
Select “Human” as your organism, and one of the classes (SNP class and/or Function class) you are looking from the limits boxes, and then press the “Go” button at the top of the page.
- 4.
You will see the total number of the class you selected in the tabs at the top of the results page.
(11/17/08)
dbSNP Growth Rate Statistics
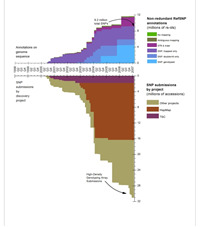
Can you show me the overall growth of dbSNP over the years since its inception?
Below is a graph of the growth of dbSNP human through February, 2007. At that point we were closing on 12 million non redundant variations clustered over 30 million submissions.
(2/27/07)
I need the growth rate for each table of dbSNP, and the size of each table in the current build of dbSNP in terms in terms of bytes.
dbSNP is now composed of a set of databases for different organisms, so your answer will be dependent upon which organism database you are interested in.
The growth rate of dbSNP depends largely on the number of submissions we receive over a given period of time — a variable which is unpredictable. We find, however, that submissions to dbSNP tend to come in bursts as we have a few submitters who submit large masses of data at one time. Such a large submission could double the size of dbSNP in a week.
Tables relating to our submissions (e.g. SubSNP, SNP, etc.) have growth rates that parallel the submission growth rate. Tables such as SnpClassCode, SnpFunctionCode rarely increase in size.(2/13/06)
Can you give me statistics on the past and expected growth of dbSNP so I can allocate an appropriate amount of disk space for the next 12 to 18 months?
dbSNP size growth has been exponential in the past, roughly doubling every year. Future growth of dbSNP will depend on what the big submitters are preparing in their submission pipeline. I am guessing there will be more non-human SNPs coming as well as more human individual genotypes and haplotype data. I would venture to say that dbSNP may grow 2 to 3 times its current size in the next 12 to 18 months. (3/8/05)
- dbSNP Data Statistics - SNP FAQ ArchivedbSNP Data Statistics - SNP FAQ Archive
Your browsing activity is empty.
Activity recording is turned off.
See more...