NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
LinkOut Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2006-.
LinkOut is a feature of NCBI databases where third parties provide information to link specific NCBI database records to relevant Web-accessible online resources, such as full-text publications, molecular biology databases (i.e., organism-specific, taxonomy, structure, etc.), catalogs of research materials (clones, cell cultures, primers, etc.), funding sources, medical resources, research groups, and others. This document explains how providers of resources other than online full text can participate in LinkOut by supplying NCBI with the necessary information for creating links from NCBI database records to the providers' resources.
How It Works
LinkOut provides links from PubMed records and other NCBI database records to online resources external to the NCBI systems. All linking information is submitted by LinkOut providers - the owner or agent for the owner of the online resource. LinkOut providers are responsible for maintaining their links.
To submit links to your resource, you will need to upload two XML files, an identity file and a resource file (see below File Preparation). The identity file contains the information about your organization needed to list your resource in LinkOut. The resource file describes the database records you will link from and contains the information that LinkOut needs to generate URLs.
Prerequisites for Participation
Resources submitted for inclusion in LinkOut will be evaluated individually to determine whether they meet our inclusion criteria.
Resources eligible for linking from NCBI databases must be directly relevant to the specific subjects of the NCBI database records and useful to users' study and research. Resources from professional societies, government agencies, educational institutions, or individuals and organizations that have received grants from major funding organizations are preferred.
Please review the Guidelines for Evaluation of Resources before applying for inclusion in LinkOut. Resources with a commercial interest should pay particular attention to the Additional Information for Commercial Interests section of the Guidelines.
Apply for Inclusion in LinkOut
To apply for inclusion in LinkOut, send an email to vog.hin.mln.ibcn@tuoknil. Include the following information:
- Name, email address, and phone number of a contact person in your organization.
- The scope of your resource, including the URL of the resource. If a username and password are required to access the resource, please include a temporary username and password that the LinkOut team can use to evaluate the resource. Also, please describe the type of NCBI database records to which you would like to apply links and provide a couple URL examples of your database records and their corresponding NCBI database records.
- Describe any restrictions on access to the resource.
File Preparation: Identity File
The identity file contains the information needed to list a provider in LinkOut. This file must be named providerinfo.xml; the file name is case sensitive. This file should be composed in a text editor, such as NotePad, not in a word processing program.
The following is an example providerinfo.xml file for the LinkOut participant, WebDatabase Co., with Provider Id 7777 and NameAbbr WebDB.
<?xml version="1.0"?>
<!DOCTYPE Provider PUBLIC "-//NLM//DTD LinkOut 1.0//EN"
"https://www.ncbi.nlm.nih.gov/projects/linkout/doc/LinkOut.dtd">
<Provider>
<!-- ProviderId is assigned by NCBI -->
<ProviderId>7777</ProviderId>
<Name>WebDatabase Co.</Name>
<NameAbbr>WebDB</NameAbbr>
<SubjectType> gene/protein/disease-specific</SubjectType>
<Attribute>registration required</Attribute>
<!-- Url is used in My NCBI and in the lists of LinkOut Providers -->
<Url>http://www.webdatabase.com</Url>
<!-- Brief is used in My NCBI -->
<Brief> On-line publisher of biomedical databases</Brief>
</Provider>
<SubjectType> and <Attribute> elements included in the providerinfo.xml file will apply to all links submitted by the provider. In the example above, access to all databases published by WebDatabase Co. requires a free registration, therefore <Attribute>registration required</Attribute> has been included in the providerinfo.xml file.
File Preparation: Resource File (XML)
Section Contents:
The resource file describes the database records to which your links will be applied and contains the information that LinkOut needs to generate URLs. Links described in the resource file must link directly to the relevant resource, requiring no additional searching to access the resource after a user clicks on the provider’s link.
XML: Resource File Format
XML resource files must have a file extension .xml; the file extension is case sensitive. File names may contain alpha-numeric characters and underscores only. Special characters and spaces are not allowed. Typically, files are named resources.xml. To help with file management, a provider may supply more than one resource file. File size may not exceed 20 MB. This file should be composed in a text editor, such as NotePad, not in a word processing program.
The resource file below describes links from NCBI’s Nucleotide database to a C. elegans sequence database provided by WebDatabase Co., ProviderId 7777.
<?xml version="1.0"?>
<!DOCTYPE LinkSet PUBLIC "-//NLM//DTD LinkOut 1.0//EN"
"https://www.ncbi.nlm.nih.gov/projects/linkout/doc/LinkOut.dtd"
[ <!ENTITY icon.url "https://www.webdatabase.com/images/webdb.gif">
<!ENTITY base.url "https://www.webdatabase.com/cgi-bin/elegans?">]>
<LinkSet>
<Link>
<LinkId>1</LinkId>
<ProviderId>7777</ProviderId>
<ObjectSelector>
<Database>Nucleotide</Database>
<ObjectList>
<Query>Caenorhabditis elegans [orgn]</Query>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>an_lookup=&lo.pacc;</Rule>
<UrlName>Caenorhabditis elegans</UrlName>
<SubjectType>organism-specific</SubjectType>
</ObjectUrl>
</Link>
</LinkSet>
<ObjectList>: Selecting Records in a Resource File
The <ObjectList> element is used to select the database records to which links will be applied. <ObjectList> contains one or more <Query> elements OR one or more <ObjId> elements. <Query> elements contain a valid search query with a valid field descriptor that will retrieve the records to which the link described in <ObjectUrl> will be applied. <ObjId> elements contain the Unique Identifier of the database records to which the link described in <ObjectUrl> will be applied.
Selecting Records Using <ObjId>
<ObjId> contains the unique identifier for a record in an NCBI database. For example, Taxonomy ID for the Taxonomy database.
| Example: Select record with Taxonomy ID 37572 in the Taxonomy database |
|---|
| <ObjectList> <ObjId>37572</ObjId> </ObjectList> |
More than one <ObjId> can be used in an <ObjectList>.
| Example: Select records with Taxonomy IDs 37572 and 33392 |
|---|
| <ObjectList> <ObjId>37572</ObjId> <ObjId>33392</ObjId> </ObjectList> |
Because <ObjId> requires more maintenance than <Query>, NCBI recommends using <Query> whenever possible. When <ObjId> is used, the provider is responsible for updating the holdings file as new records are submitted to PubMed.
Selecting Records Using <Query>
The <Query> element contains a valid NCBI database search. A valid search term or terms should include the corresponding field descriptors. For example, a search query for Arabidopsis Thaliana would be: Arabidopsis Thaliana[orgn]. A search query for the Genbank accession number HM047434 would be HM047434 [pacc].
See Entrez Help for information on constructing search queries and field descriptors. Links will be applied to the citations retrieved by the search.
Tips for Using <Query>
- 1.
Ranging is not allowed in Unique Identifier searches. For journal searches, ranging is additionally not allowed in Volume, Issue, or Page searches.
- 2.
Truncation is not allowed in search statements.
- 3.
Search field descriptors (for example, [orgn] for organism or [pacc] for primary accession number) must be used with <Query>
- 4.
To include a date range in searches, use this format: startyear:endyear[dp]. Dates should be notated as YYYY/MM/DD. Month and Day are optional.
- 5.
Do not use the search field descriptors [sb] or [filter].
- 6.
Boolean operators AND, OR, NOT must be in uppercase.
- 7.
Use either NLM’s Title Abbreviations [ta] or ISSN numbers in journal searches. Title Abbreviations must be entered in double quotes, e.g., “J Mol Dis” [ta].
Example: Select records with the organism “Caenorhabditis elegans” published from 1996 to 1999 in the Nucleotide database <Database>Nucleotide</Database>
<ObjectList>
<Query>Caenorhabditis elegans [orgn] AND 1996:1999 [pdat]</Query>
</ObjectList>
See the results of this <Query> in the Nucleotide database.
| Example: Select records with the organism “Caenorhabditis elegans” published by J. Smith in Nucleotide. As new records are submitted to the database, links will be applied automatically. |
|---|
| <Database>Nucleotide</Database> <ObjectList> <Query>Caenorhabditis elegans [orgn] AND smith j [auth]</Query> </ObjectList> |
See the results of this <Query> in the Nucleotide database.
More than one <Query> can be listed within the <ObjectList>, as shown in the example below.
| Example: Select records for chimpanzees starting from the publication date January 1, 2000 and records for humans starting from January 1, 2002 in the Genome database. As new records are submitted, links will be applied automatically. |
|---|
| <Database>Genome</Database> <ObjectList> <Query>chimpanzee [orgn] AND 2000:2010[dp]</Query> <Query>human [orgn] AND 2002:2010[dp] </Query> </ObjectList> |
See the results for this <ObjectList> in the Genome database.
Additional Information on Using <Query> for Linking
When using Genbank accession numbers with <Query> include the field descriptor for primary accession number [pacc] to ensure that search results are directly related to the Genbank number included in the search query.
| Example: Use a Genbank accession number as a search query to create links in Nucleotide. |
|---|
| <Database>Nucleotide</Database> <ObjectList> <Query>HM047434 [pacc]</Query> </ObjectList> |
See the results for this <Query> in Nucleotide.
MeSH headings can be used to create links in PubMed. In this case, the <Query> should be very precise. Only Major headings should be used in the <Query> and noexp should be used so the terms will not be exploded.
| Example: Use MeSH headings to select citations on acupuncture therapy methods in PubMed. |
|---|
| <Database>PubMed</Database> <ObjectList> <Query>”Acupuncture Therapy/methods”[majr:noexp] </Query> </ObjectList> |
See the results for this <Query> in PubMed.
<ObjectUrl>: Specifying the Link to Access the Resource
The <ObjectUrl> element is used to describe the link to the online resource. <ObjectUrl> contains the sub-elements <Base>, <Rule>, <SubjectType>, <Attribute>, and <UrlName>. <Base> and <Rule> are concatenated to form the URL for the link. <SubjectType>, <Attribute>, and <UrlName> describe the resource to which the record is being linked.
Creating a URL for a Link
<Base> is the stable portion of the URL for the provider’s resource. This is usually the provider website or CGI program.
<Rule> is the remainder of the URL needed to access the appropriate record within the resource.
| Example: Create the URL https://www.webdatabase.com/cgi-bin/elegans?OID=1988 |
|---|
| <Base>https://www.webdatabase.com/ cgi-bin/elegans?<Base> <Rule>OID=1988</Rule> |
If the URL for the resource follows a pattern using variable values that are found in a database record, the pattern can be described in the <Rule> element, and LinkOut can insert the appropriate values for each citation. This allows many links to be generated from the information in a single <ObjectUrl>.
URL patterns are described using LinkOut’s XML entities. An XML entity is a short text string that represents a type of value. During LinkOut processing, the text string is replaced in the URL by the appropriate value for each record.
| Example: Create URLs following the pattern: https://www.webdatabase.com/cgi-bin/an_lookup=[PACC] |
|---|
| <Base>https://www.webdatabase.com/cgi-bin/</Base> <Rule> an_lookup=&lo.pacc;</Rule> |
Using this <Base> and <Rule>, the URL constructed for the record with accession number AL032671 would be https://www.webdatabase.com/cgi-bin/elegans?an_lookup=AL032671
Entities can be combined with other information in the <Rule>.
| Example: Create URLs following the pattern: https://www.webdatabase.com/cgi-bin/db=elegans&id_lookup=[NCBI database Unique Identifier]&view=text |
|---|
| <Base>https://www.webdatabase.com/cgi-bin</Base> <Rule>/db=elegans&id_lookup=&lo.id;&view=text</Rule> |
In this case, the URL generated for the record with the unique ID "6016240" would be: https://www.webdatabase.com/cgi-bin/db=elegans&id_lookup=6016240&view=text
LinkOut does not support Unicode (UTF-8) and requires that certain special characters be encoded in files.
Describing the Resource
The relevance of resources linked from NCBI database records should be readily apparent to users. The name and/or description of the resource should convey something about the information that is being offered and its relevance.
The elements <SubjectType> and <Attribute> are used in the <ObjectUrl> to describe resources. Available SubjectTypes can be found in Special Elements: SubjectType. Available Attributes can be found in Special Elements: Attribute.
If the available SubjectTypes and Attributes do not suffice to describe the resource, UrlName can be used as well. If no SubjectType is given, the SubjectType “miscellaneous” will be assigned automatically.
The application of SubjectTypes and Attributes is at the discretion of the resource provider. However, if there are any barriers to accessing the resource, one of the following Barrier Attributes must be used:
<Attribute>registration required</Attribute>
<Attribute>subscription/membership/fee required</Attribute>
Continuing the example above, if WebDatabase Co. requires a subscription to access the C. elegans database, the <ObjectUrl> element might look like this:
<ObjectUrl>
<Base>https://www.webdatabase.com/cgi-bin/</Base>
<Rule>/db=elegans&id_lookup=&lo.id;&view=text</Rule>
<Attribute>subscription/membership/fee required</Attribute>
</ObjectUrl>
Resource File Examples
Example 1: Molecular Biology Database, Inc., Provider Id 1234, provides links to freely available information for the Taxonomy records with IDs 9606 and 111063. URLs for the database entries are created using a text string that is not included in Taxonomy, so links are created individually for each record. To minimize the repetition of textual data, the Base for the URL has been defined as an Entity in the Prolog of the file.
<!DOCTYPE LinkSet PUBLIC "-//NLM//DTD LinkOut 1.0//EN"
"https://www.ncbi.nlm.nih.gov/projects/linkout/doc/LinkOut.dtd"
[<!ENTITY base.url "http://molbioco.com/animals/">]>
<LinkSet>
<Link>
<LinkId>1</LinkId>
<ProviderId>1234</ProviderId>
<ObjectSelector>
<Database>taxonomy</Database>
<ObjectList>
<ObjId>9606</ObjId>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>homo/h._sapiens</Rule>
<UrlName>Homo sapiens</UrlName>
<SubjectType>taxonomy/phylogenetic</SubjectType>
</ObjectUrl>
</Link>
<Link>
<LinkId>2</LinkId>
<ProviderId>1234</ProviderId>
<ObjectSelector>
<Database>taxonomy</Database>
<ObjectList>
<ObjId>9733</ObjId>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>orcinus/o._orca</Rule>
<UrlName>Orcinus orca</UrlName>
<SubjectType>taxonomy/phylogenetic</SubjectType>
</ObjectUrl>
</Link>
</LinkSet>
Example 2: Genotypes, Inc., Provider Id 4321, provides free online access to genotyping assays from records in the SNP database. SNP records are selected using the SNP unique identifier. The URL to access the assays at their site follows this pattern for each record: https://gti.com/Gateway?source= SNP&res=Assays&ap1=rs[SNP ID]
<?xml version="1.0"?>
<!DOCTYPE LinkSet PUBLIC "-//NLM//DTD LinkOut 1.0//EN"
"https://www.ncbi.nlm.nih.gov/projects/linkout/doc/LinkOut.dtd"
[<!ENTITY base.url
"https://gti.com/Gateway?source=SNP&res=Assays&">]>
<LinkSet>
<Link>
<LinkId>1</LinkId>
<ProviderId>4321</ProviderId>
<ObjectSelector>
<Database>SNP</Database>
<ObjectList>
<ObjId>7928656</ObjId>
<ObjId>2049045</ObjId>
<ObjId>1811350</ObjId>
<ObjId>1871598</ObjId>
<ObjId>7947824</ObjId>
<ObjId>681267</ObjId>
<ObjId>1947741</ObjId>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>ap1=rs&lo.id;</Rule>
<UrlName>Genotyping Assays</UrlName>
</ObjectUrl>
</Link>
</LinkSet>
Example 3: A record may be retrieved by more than one <Query>. When this happens, link assignment will be handled as described in the LinkOut Policy: Duplicate Links and Multiple Links.
If these queries are in different Link elements, <Attribute>preference</Attribute> can be used to indicate which <Link> element should be applied to the record. This is generally used in situations where the links for a subset of a range have a different URL pattern or different access restrictions. In the example below, the records included in LinkId 1 will also be selected by LinkId 2.
The LinkOut provider WebDatabase Co. provides links from the Nucleotide database to the C. elegans sequence database.
LinkId 1 describes links from particular Nucleotide records. The records are selected using <Query> and have a special <Rule>. Because these records are also included in LinkId 2, <Attribute>preference</Attribute> is used to indicate that only these links should be applied to these citations.
LinkId 2 provides links from all Nucleotide records on C. elegans to WebDatabase Co.’s C. elegans records, except for the records selected in LinkId 1.
As LinkId 1 describes specific requirements, it is listed before the general LinkId 2.
<?xml version="1.0"?>
<!DOCTYPE LinkSet PUBLIC "-//NLM//DTD LinkOut 1.0//EN"
"https://www.ncbi.nlm.nih.gov/projects/linkout/doc/LinkOut.dtd"
[<!ENTITY base.url "https://www.webdatabase.com/cgi-bin/elegans?">]>
<LinkSet>
<Link>
<LinkId>1</LinkId>
<ProviderId>7777</ProviderId>
<ObjectSelector>
<Database>Nucleotide</Database>
<ObjectList>
<Query>Caenorhabditis elegans [orgn] AND 1997:1999 [pdat] AND smith j [auth]</Query>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>auth_lookup=j-smith&view=pdf</Rule>
<Attribute>Full-text PDF</Attribute>
<Attribute>preference</Attribute>
</ObjectUrl>
</Link>
<Link>
<LinkId>2</LinkId>
<ProviderId>7777</ProviderId>
<ObjectSelector>
<Database>Nucleotide</Database>
<ObjectList>
<Query>Caenorhabditis elegans [orgn]</Query>
</ObjectList>
</ObjectSelector>
<ObjectUrl>
<Base>&base.url;</Base>
<Rule>an_lookup=&lo.pacc;&view=full</Rule>
</ObjectUrl>
</Link>
</LinkSet>
File Preparation: Resource CSV File
Links data can also be provided in CSV (comma separated values) files. The CSV resource file contains LinkOut data provider identifiers, database record Ids or queries, and links data to your resource pages, all of which is used to create links in NCBI databases.
A LinkOut program converts CSV files in to XML files that validate against the LinkOut DTD. Links provided in CSV files must link directly from a NCBI database record to a resource page that provides information directly related to the database record.
CSV files need to have the file extension .csv; the file extension is case sensitive. File names may contain alpha-numeric characters and underscores only. Special characters and spaces are not allowed. Examples of file name and extension: linksgene2015.csv, or linksnucleotide2015.csv. To help with file management, a provider may submit more than one resource file. CSV files may not exceed 10 MB each.
Section Contents
Resource CSV File Data Fields
The CSV files used by LinkOut to create links have required and optional data fields:
Field 1: PrId (required). Provider Id assigned by NCBI to links data providers. A four-digit number.
Field 2: DB (required). NCBI database name. Enter the name of the NCBI database for which you want to provide links data.
Field 3: UID or Query (required). Each record in an NCBI database has a numerical unique identifier (UID). For example, in this Taxonomy record: https://www.ncbi.nlm.nih.gov/taxonomy/?term=37572 the Taxonomy Id 37572.
NCBI database records can also be retrieved using queries. For example, Nucleotide query: Caenorhabditis elegans [orgn] AND 2011:2015[pdat] AND smith j [auth]
Field 4: URL (required). URL to the supplemental information page in your resource, which is directly related to the selected NCBI database record.
Filed 5: IconUrl (optional). URL of an icon file that you would like to represent your resource. The icon should meet the specifications described in Icons. The icon URL should point directly to the icon file in your server. If an icon is not provided, LinkOut will use the LinkOut generic icon. Icons are only displayed in PubMed.
Field 6: UrlName(optional). Additional description about the link to content.
Field 7: SubjectType (required*). In this field enter a subject type selected from this page that best describes your resource. (*) If the subject type is present in the identity file, this field should be left empty.
Field 8: Attribute (required). If access to the resource requires a license or membership, enter the following in this field: Subscription/membership/fee required. If access is free, but registration is required enter: registration required. Otherwise, if access to the resource is free and registration is not required, this field can be left empty.
Resource File CSV Format
Your CSV file can be formatted as a table. Each field must be separated by commas.
Field 1: PrId. Provider Id, a four-digit number. For example: 1234.
Field 2: DB. Enter the selected NCBI database name in this field. For example: Nucleotide
Field 3: Two options UID or Query:
UID. Taxonomy ID. For example: 37572
Query. A query that retrieves the Nucleotide records selected: Caenorhabditis elegans [orgn] AND 2011:2015[pdat] AND smith j [auth]
Field 4: URL. https://treebase.org/treebase-web/search/taxonSearch.html
Filed 5: IconUrl. Only needed for PubMed. Leave blank for other databases.
Field 6: UrlName. Caenorhabditis elegans
Field 7: SubjectType. organism-specific (*) If the ‘organism-specific’ subject type is present in the identity file, this field should be left blank.
Field 8: Attribute. If access to the resource requires a license or membership, enter the following in this field: Subscription/membership/fee required. If access is free, but registration is required enter: registration required. Otherwise, if access to the resource is free and registration is not required, this field can be left empty.
Resource CSV File Examples
Example 1. Access to a resource is through license or membership: Provider Id 1234, provides links for the Nucleotide record 3810674. The URL provided leads to a page that has information that supplements the Nucleotide record. The UrlName indicates the resource page topic. The subject type was provided in the identity file, and consequently this field is left blank. Access to the resource is through license or membership only, therefore enter the attribute “subscription/membership/fee required” in this field.
Note that each field must be comma separated.
Field 1: PrId. 1234
Field 2: DB. Nucleotide
Field 3: Query. BX284601.5[pacc]
Field 4: URL. http://www.origene.com/cdna/search-all.mspx?product=HCLONES&term=1B%20%28VP2%29
Filed 5: IconUrl. None needed for Nucleotide.
Field 6: UrlName Caenorhabditis elegans chromosome I
Field 7: SubjectType.
Field 8: Attribute. Subscription/membership/fee required
A sample file using a spreadsheet program such as MS Excel.
| PrId | DB | Query | URL | IconUrl | UrlName | SubjectType | Attribute |
|---|---|---|---|---|---|---|---|
| 1234 | Nucleotide |
| http://www.origene.com/cdna/search-all.mspx?product=HCLONES&term=1B(VP2) |
| Subscription/membership/fee required |
The same entries in a sample CSV file can be downloaded here. Save files with the extension .csv, and upload them to the “holdings” directory of the FTP assigned to you.
Example 2. Access to a resource is free, but requires registration: Provider Id 1234, provides links for Gene database records, however, queries are used instead of UID numbers. The URL provided leads to a page that has information that supplements the Gene record. The UrlName field is left blank. The subject type was not provided in the identity file, and consequently it must be listed here. Access to the resource is free, but registration is required, therefore, enter the attribute “registration required” in this field. Note that each field must be comma separated.
Field 1: PrId. 1234
Field 2: DB. Gene
Field 3: Query. APOE[sym] AND chromosome 7
Field 4: URL. https://biogps.org/#goto=genereport&id=11816
Filed 5: IconUrl. None needed for Gene
Field 6: UrlName
Field 7: SubjectType. gene/protein/disease-specific
Field 8: Attribute. Registration required
A sample file using a spreadsheet program such as MS Excel.
| PrId | DB | Query | URL | IconUrl | UrlName | SubjectType | Attribute |
|---|---|---|---|---|---|---|---|
| 1234 | Gene | APOE[sym] AND chromosome 7 | https://biogps.org/#goto=genereport&id=11816 | gene/protein/disease-specific | Registration required |
The same entries in a sample CSV file can be downloaded here. Save files with the extension .csv, and upload them to the “holdings” directory of the FTP assigned to you.
Example 3. Access to a resource is free and registration is not required: Provider Id 1234, provides links for the Taxonomy record 314297. The URL provided leads to a page that has information that supplements the Taxonomy record. The UrlName indicates the resource page topic. The subject type was provided in the identity file, and consequently this field is left blank. Access to the resource is free and registration is not required, consequently, the field is left blank. Note that each field must be comma separated.
Field 1: PrId. 1234
Field 2: DB. Taxonomy
Field 3: UID. 314297
Field 4: URL. https://www.marinespecies.org/aphia.php?p=taxdetails&id=611463
Filed 5: IconUrl. None needed for Taxonomy
Field 6: UrlName Compsopogon hookeri Montagne
Field 7: SubjectType.
Field 8: Attribute.
A sample file using a spreadsheet program such as MS Excel.
| PrId | DB | UID | URL | IconUrl | UrlName | SubjectType | Attribute |
|---|---|---|---|---|---|---|---|
| 1234 | Taxonomy | 314297 | https://www.marinespecies.org/aphia.php?p=taxdetails&id=611463 | Compsopogon hookeri Montagne |
The same entries in a sample CSV file can be downloaded here. Save files with the extension .csv, and upload them to the “holdings” directory of the FTP assigned to you.
File Preparation: Resource File (Simple Text)
Section Contents:
Simple Text: Resource File Format
Providers may choose to submit the resource file in a simple text file instead of XML.
Text resource files must have a file extension .ft; the file extension is case sensitive. File names may contain alpha-numeric characters and underscores only. Special characters and spaces are not allowed. Typically, files are named “resources.ft”. To help with file management, a provider may supply more than one resource file. File size may not exceed 10 MB.
This file should be composed in a text editor, such as NotePad, not in a word processing program.
The resource file below describes links from NCBI’s Nucleotide database to a C. elegans sequence database provided by WebDatabase Co., ProviderId 7777.
--- lines starting with "-" are comments ---
--- information in the first block is global ---
prid: 7777
dbase: nucleotide
stype: organism-specific
!base: http://www.webdatabase.com/cgi-bin/elegans?
------
linkid: 1
query: Caenorhabditis elegans [orgn]
base: &base;
rule: an_lookup=&lo.pacc;
name: Caenorhabditis elegans
------
Simple Text: Global Information
The first block holds global information that will be used throughout the file.
prid: LinkOut Provider ID
dbase: NCBI database that will be hosting the links, e.g., pubmed, nucleotide, taxonomy
stype: SubjectType. See Special Elements: SubjectType for all available SubjectTypes.
attr: Attribute. See Special Elements: Attribute for all available Attributes.
Simple Text: Creating Links
Each subsequent block specifies a LinkOut link. This has two basic parts, a valid search query with a valid field descriptor and a URL pointing back to the provider's site. Each search query will be evaluated, and a link to the specified URL will be applied to records that are retrieved by the query. See Entrez Help for information on constructing search queries and on field descriptors. Links will be applied to the citations retrieved by the search.
In the simplest case, each block could be:
-----
query: [a valid NCBI database query with a valid database field descriptor]
rule: [the URL that will be applied to the records retrieved by the query]
-----
Simple Text: Selecting Records
Any valid search query may be used to select records. Each query should appear on a single query line and must include a database field descriptor. Multiple query lines in one block will be OR-ed together: See Entrez Help for information on constructing search queries and on field descriptors.
| Example: This search will be translated as: human[name] OR chimpanzee[name] |
|---|
| query: human [name] query: chimpanzee [name] |
Genbank accession numbers can be used as queries to create links. For example: a Genbank sequence accession number for Arabidopsis Thaliana is HM047434. A query for this sequence would be HM047434 [pacc] – pacc is the field descriptor for primary accession numbers.
| Example: enter the query with the field descriptor [pacc] |
|---|
| HM047434 [pacc] |
Each record in an NCBI database has a numerical unique identifier (UID). You can select the NCBI database records that you would like to link from by UID in the uids: line.
| Example: Place links on records with UIDs 123456, 123469, and 3847559 |
|---|
| ---- separate unique identifiers (UIDs) with a blank space. Each new line should start with the “uids:” label --- uids: 123456 123469 3847559 |
Simple Text: Specifying the Link
The link is specified using the base: and rule: lines. base: is the stable portion of the URL for the resource. This is usually the URL of the provider’s website or CGI program. rule: is the remainder of the URL needed to access the online resource.
base: and rule: are concatenated to form the URL for the link.
If desired, the entire URL for the resource can be included on the rule: line, and the base: line can be omitted.
| Example: The following will both create a link to the URL http://www.webdatabase.com/cgi-bin/elegans?OID=1988 |
|---|
| ------ rule: https://www.webdatabase.com/ cgi-bin/elegans?OID=1988 ------- ------ base: https://www.webdatabase.com/cgi-bin/elegans? rule: OID=1988 ------ |
If the URL for the resource follows a pattern using variable values that are found in the database record, the pattern can be described on the rule: line, and LinkOut can insert the appropriate values for each citation.
URL patterns are described using LinkOut’s XML entities. An XML entity is a short text string that represents a type of value. During LinkOut processing, the text string is replaced in the URL by the appropriate value for each record.
| Example: Create URLs following the pattern: http://www.webdatabase.com/cgi-bin/an_lookup=[PACC] |
|---|
| base: https://www.webdatabase.com/cgi-bin/ rule: an_lookup=&lo.pacc; |
Using this base: and rule:, the URL constructed for the record with accession number AL032671 would be https://www.webdatabase.com/cgi-bin/elegans?an_lookup=AL032671
Entities can be combined with other information in the rule:
| Example: Create URLs following the pattern: https://www.webdatabase.com/cgi-bin/db=elegans&id_lookup=[NCBI database Unique Identifier]&view=text |
|---|
| base: https://www.webdatabase.com/cgi-bin rule: db=elegans&id_lookup=&lo.id;&view=text |
In this case, the URL generated for the record with the unique ID 6016240 would be: https://www.webdatabase.com/cgi-bin/db=elegans&id_lookup=6016240&view=text
To minimize the repetition of textual data, the base: portion of the URL can be defined as an entity in the global information block, as shown below.
prid: 4592
dbase: PubMed
!base.url: https://a257.g.akamaitech.net/7/257/2422/
-------------------------------------------------------------------
linkid: 704411419
uids: 15754467
base: &base.url;
rule: 01jan20051800/edocket.access.gpo.gov/2005/pdf/05-4062.pdf
attr: full-text PDF
-------------------------------------------------------------------
linkid: 70389516
uids: 15736310
base: &base.url;
rule: 01jan20051800/edocket.access.gpo.gov/2005/pdf/05-3829.pdf
attr: full-text PDF
-------------------------------------------------------------------
linkid: 70379232
uids: 15732197
base: &base.url;
rule: 01jan20051800/edocket.access.gpo.gov/2005/pdf/05-3728.pdf
attr: full-text PDF
-------------------------------------------------------------------
Simple Text: Describing the Resource
The relevance of resources linked from NCBI database records should be readily apparent to users. The name and/or description of the resource should convey something about the information that is being offered and its relevance.
The following optional fields allow you to describe your links and resources.
icon: URL of an icon file that you would like to represent your link and resources. Only applicable to links in PubMed. The icon should meet the specifications described in Icons.
name: Additional description of the link. name: should only be used when the values in the LinkOut SubjectType and Attribute lists do not suffice.
stype: SubjectType. See Special Elements: SubjectType for all available SubjectTypes. SubjectType is used to determine where links will be placed in the LinkOut Display. If no SubjectType is given, the SubjectType “miscellaneous” will be assigned automatically.
attr: Attribute. See Special Elements: Attribute for all available Attributes.
The application of SubjectTypes and Attributes is at the discretion of the resource provider. However, if there are any barriers to accessing the resource, one of the following Barrier Attributes must be used:
registration required
subscription/membership/fee required
Simple Text: Resource File Examples
Example 1: The following file shows five links to taxonomic resource on the Web. Because each link has an individual URL, the links are made separately.
----------NCBI taxonomy bookmark links ----
prid: 3206
dbase: Taxonomy
------Apis mellifera)------
linkid: 1
query: Apis mellifera [name]
rule: http://beelab.cas.psu.edu/intro.html
name: Honey Bee Lab (Penn State)
------
linkid: 2
query: Apis mellifera [name]
rule: http://www.barc.usda.gov/psi/brl/
name: Bee Research Lab (USDA Beltsville)
------
linkid: 3
query: Apis mellifera [name]
rule: http://www.hgsc.bcm.tmc.edu/projects/honeybee/
name: Baylor Honey Bee Genome
------
linkid: 4
query: Apis mellifera [name]
rule: http://titan.biotec.uiuc.edu/bee/honeybee_project.htm
name: Honey Bee Brain EST Project
------
linkid: 5
query: Apis mellifera [name]
rule: http://ourworld.compuserve.com/homepages/Beekeeping/weblinks.htm
name: Bee Web Links
------
Example 2: The hypothetical provider Genotypes, Inc., Provider Id 4321, provides free online access to genotyping assays from records in the SNP database. SNP records are selected using the SNP unique identifier. The URL to access assays at their site follows this pattern for each record: https://genotypinc.com/servlet/web.Gateway?source=NCBI.SNP&res=genotypAssay&ap1=rs[SNP ID]
-----Geotypes SNP links global info ---
prid: 4321
dbase: SNP
-----Begin Links ---
linkid: 1
uids: 7928656 2049045 1811350 1871598 7947824 681267 1947741
base: https://genotypinc.com/servlet/web.Gateway?
source=NCBI.SNP&res=genotypAssay&
rule: ap1=rs&lo.id;
name: Genotyping Assays
------
Example 3:
A record may be retrieved by more than one <Query>. When this happens, link assignment will be handled as described in Duplicate Links and Multiple Links.
If these queries are in different linkids, you can use attr: preference to indicate which link should be applied to the record. This is generally used in situations where the links for a subset of a range have a different URL pattern or different access restrictions.
In the example below, the records included in LinkId 1 below will also be selected by LinkId 2.
The hypothetical LinkOut provider WebDatabase Co. provides links from the Nucleotide database to the C. elegans sequence database.
LinkId 1 describes links from all Nucleotide records on C. elegans published by J. Smith from 1997 to 1999 to a set of C. elegans records in PDF format. Because these records are also included in LinkId 2, attr: preference is used to indicate that only this link should be applied to these citations.
LinkId 2 provides links from all Nucleotide records on C. elegans to WebDatabase Co.’s C. elegans records, except for the records selected in LinkId 1.
Because LinkId 1 describes specific requirements, it is listed before the general LinkId 2.
----- Nucleotide links ----
prid: 7777
dbase: nucleotide
!base: "https://www.webdatabase.com/cgi-bin/elegans?"
------
linkid: 1
query: Caenorhabditis elegans [orgn] AND
1997:1999 [pdat] AND smith j [auth]
base: &base;
rule: auth_lookup=j-smith&view=pdf
attr: full-text PDF
attr: preference
------
linkid: 2
query: Caenorhabditis elegans [orgn]
base: &base;
rule: an_lookup=&lo.pacc;&view=full
------
Example 4: The following file shows three links to The Restriction Enzyme Database. Each query uses a specific sequence accession number. Because each link has an individual URL, the links are made separately.
prid: 1234
dbase: Nucleotide
!base: https://rebase.neb.com/rebase/enz/
------------
linkid: 1
query: U65398[pacc]
base: &base;
rule: 7.html
name: REBASE enzyme AatII
------
linkid: 2
query: X62690[pacc]
base: &base;
rule: 8.html
name: REBASE enzyme AbrI
------
linkid: 3
query: D10671 [pacc]
base: &base;
rule: 18.html
name: REBASE enzyme AccI
------
File Evaluation
After your application for inclusion in LinkOut has been accepted, prepare an identity file and sample resource files according to the instructions above. Resource files should contain links to at least five records from the selected database.
Validate the files using the LinkOut File Validation Utility. Email the files to vog.hin.mln.ibcn@tuoknil.
Your files will be evaluated by the LinkOut team, and you will be contacted regarding any corrections. The evaluation process will continue until your files are substantially error free.
Account Assignment
When the submitted files are substantially error free, you will be assigned a ProviderId (PrId) and an approved name abbreviation (NameAbbr), and you will be given a password for an NCBI private FTP account.
Please note that each provider will be given only one account at NCBI.
File Transfer
When you receive your account information, validate the files using the LinkOut File Validation Utility and transfer all files via FTP to the host FTP-private.ncbi.nlm.nih.gov. Place the files in the “holdings” directory of your FTP account. No subdirectories may be created in the holdings subdirectory.
When files have been submitted, inform the LinkOut team at vog.hin.mln.ibcn@tuoknil. Your files will be given a final evaluation before being placed in the production queue. From this point on, files will be processed automatically every day.
Links should appear in the selected NCBI database within 2 days of file submission. If 2 days have passed and you do not see your links, please write to vog.hin.mln.ibcn@tuoknil.
File Maintenance
Provider Responsibilities
Link providers are responsible for:
- maintaining their LinkOut files
- transferring any additions, changes or deletions of their links to NCBI
- updating files and informing NCBI when access rights are changed
- correcting broken or incorrect links in a timely manner
Providers may transfer new versions of current files or add new resource files at any time. It is the responsibility of the provider to keep files current and valid. Links are regenerated every day based on the resource files in each provider’s directory. Therefore, providers must delete obsolete files from their /holdings directory.
Additional provider responsibilities are described in LinkOut Policies: Provider Responsibilities.
Confirmation and Error Messages
Upon processing an updated file, NCBI will send an acknowledgment to the designated LinkOut contact. If you prefer not to receive this acknowledgment, please notify the LinkOut Team.
If files cannot be processed because of errors, a message with the subject line “LinkOut files uploaded to NCBI - Critical ERRORS!” will be sent to the LinkOut contact. In this case, please correct the files and resubmit them. If you have any questions about the errors, contact LinkOut at vog.hin.mln.ibcn@tuoknil.
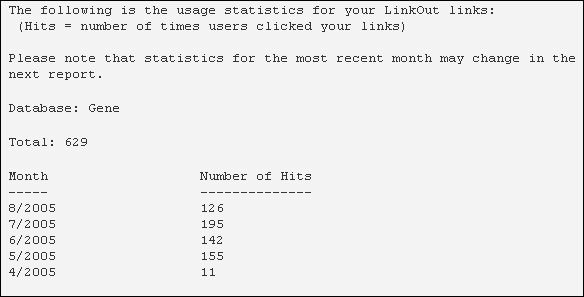
Provider Statistics
LinkOut collects statistics on the number of clicks on each providers’ links in the LinkOut display.
Statistics can be emailed to the LinkOut contact monthly. If you would like to receive statistics, please notify the LinkOut Team at vog.hin.mln.ibcn@tuoknil
Statistics send via email include the yearly and monthly totals for clicks on a provider’s links (a CSV file with the same information is provided as an attachment as well).
Statistics may change for the first 2 weeks that they are available. After 2 weeks, statistics will be stable.
A sample statistics report is shown below.
- How It Works
- Prerequisites for Participation
- Apply for Inclusion in LinkOut
- File Preparation: Identity File
- File Preparation: Resource File (XML)
- File Preparation: Resource CSV File
- File Preparation: Resource File (Simple Text)
- File Evaluation
- Account Assignment
- File Transfer
- File Maintenance
- Provider Statistics
- Information for Other Resource Providers - LinkOut HelpInformation for Other Resource Providers - LinkOut Help
Your browsing activity is empty.
Activity recording is turned off.
See more...
