BLAST is a Registered Trademark of the National Library of Medicine
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
BLAST® Command Line Applications User Manual [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2008-.
BLAST supports two different methods to multi-thread your search. Depending on the situation, one method may be faster than another. Starting with the 2.15.0 release, BLAST will select your fastest method. Both methods produce the same results. This section describes the methods, how BLAST selects one, and how you can override the automatic selection (not recommended).
The two methods are:
- ThreadByQuery: This method works well for input with many queries and a (relatively) small database. With this method, every thread receives a batch of queries, searches the entire database, and formats the results. The thread will then go back to see if more queries need to be searched. Whether this method is faster depends on the program (e.g., BLASTP or BLASTN) and the task (e.g., blastp-fast). See Table 1-Minimum Sizes for FASTA Query below for details.
- ThreadByDatabase: This method works well for larger databases and handles any number of queries well. With this method, all threads process one batch of queries, splitting up the work of searching the database.
BLAST now automatically selects the best threading model based on the table data below. Limiting your search by taxid (using -taxids or -taxidlist) or by a GI list effectively decreases your database size. BLAST will consider that information when deciding on the best threading model.
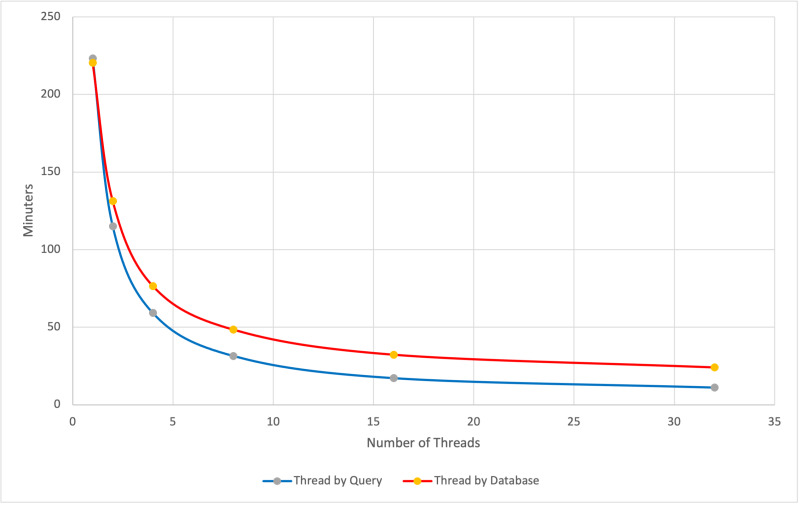
Below, we show an example where ThreadByQuery works well. The orange line shows results for ThreadByQuery, where each thread works independently on a batch of sequences. The blue line shows the ThreadByDatabase method appropriate for large databases or a few queries.
BLASTP search of 19,338 human proteins from the refseq_select protein database against swissprot. The top (red) line is ThreadByDatabase, and the bottom (blue) line is ThreadbyQuery. For 32 threads, the search is 2.2 times faster (11 minutes vs 24 minutes) with ThreadingByQuery. The task blastp-fast was used.
Though this is not recommended, you may override the automatic selection by BLAST with the -mt_mode. Use one as a value for ThreadByQuery and 2 for ThreadByDatabase. The default value for the -mt_mode option is 0, which means BLAST selects a method for you. You can specify the number of threads with the “-num_threads” option. See the command line example below.
$ blastp –db swissprot -query BIGFASTA.fsa –out test.out -num_threads 32 -mt_mode 1
To run ThreadByQuery well, the query file should be above a minimum size and the database below a maximum size. Minimum sizes (in bytes) for FASTA query files and maximum database sizes (bases or residues) are listed in the table below. Both criteria must be satisfied for BLAST to run a search with ThreadByQuery. We established the values in this table through a large number of experiments and have found them to be reliable in almost all cases.
Table 1
-Minimum Sizes for FASTA Query
| Program | Task | Query file size (bytes) | Database (letters) |
|---|---|---|---|
| BLASTN | megablast | 10,000,000 | 6,000,000,000 |
| BLASTN | blastn | 1,500,000 | 150,000,000 |
| BLASTP | blastp-fast | 200,000 | 2,500,000,000 |
| BLASTP | blastp | 200,000 | 740,000,000 |
| BLASTX | blastx-fast | 1,250,000 | 900,000,000 |
| BLASTX | blastx | 1,250,000 | 900,000,000 |
| TBLASTN | tblastn-fast | 200,000 | 350,000,000 |
| TBLASTN | tblastn | 200,000 | 280,000,000 |
- Threading By Query - BLAST® Command Line Applications User ManualThreading By Query - BLAST® Command Line Applications User Manual
Your browsing activity is empty.
Activity recording is turned off.
See more...