BLAST is a Registered Trademark of the National Library of Medicine
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
BLAST® Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2008-.
Introduction
In addition to providing BLAST sequence alignment services on the web, NCBI also makes these sequence alignment utilities available for download through FTP. This allows BLAST searches to be performed on local platforms against databases downloaded from NCBI or created locally. These utilities run through DOS-like command windows and accept input through text-based command line switches. There is no graphic user interface.
The following tutorial discusses the steps needed to install BLAST+ and a sample NCBI database on PCs running Windows 10 Operating System.
Downloading
The BLAST+ software package is available as self-extracting archives. The archive ncbi-blast-#.#.#+-win64.exe, is for PCs running 64-bit Windows operating system. Here, the "#.#.#" denotes the current version number of the package. Archives with the same base name and version number are equivalent.
Please note that the archive with the ".tar.gz" file extension does not have the installer function. The discussion below focuses on archives with the ".exe" extension.
Steps
Steps to download the package are:
- Point a browser to this FTP directory:
https://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/ - Right click on a desired archive and select "Save target as…" from the popup menu
- In the prompt, switch to a desired directory (folder) and click the "Save" button to save the archive to the selected location on the local disk
Examples
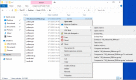
These steps for the "ncbi-blast-2.10.0+-win64.exe" archive are given in Figure 1a and Figure 1b, where the first two steps are demonstrated by 1a and the last step is demonstrated by 1b.
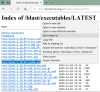
Figure 1a.
Download a blast+ package from NCBI through a web browser: Log on to ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/ and select "Save target as ..." after right-clicking on "ncbi-blast-2.2.29+-win64.exe".
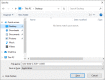
Figure 1b.
Download a blast+ package from NCBI through a web browser: Change the location in the subsequent prompt to your own directory under "C:" before saving the archive to a desired location.
Installation
The BLAST+ archive downloaded above contains a built-in installer. Double click the file to launch the installer, accept the license agreement to specify the install location in a new prompt. In this test case "C:\Users\taota\Desktop\blast-2.10.0+" is set as the installation directory (Figure 2). Clicking the "Install" button, the installer will create this directory and install the following:
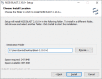
Figure 2.
Windows BLAST+ installer dialog box: Use “Browse…” button to change the name and location of blast+ installation.
- a "doc" subdirectory with the BLAST+ user manual in PDF format
- an "uninstaller" for future removal of the installation, and
- a "bin" subdirectory with all BLAST programs and accessory utilities
Table 1 sums up programs and utilities provided by the BLAST+ package.
Table 1
Programs and utilities contained in the blast+ package
| Program | Function |
|---|---|
| blastdbcheck | Examines random entries in the target BLAST database to confirm its integrity |
| blastdbcmd | Retrieves sequences or other information from an existing BLAST database. Specific sequence retrieval requires the database be created with -parse_seqids option |
| blastdb_aliastool | Creates database alias to tie several databases together for example or to specify a subset of sequence in a target database |
| blastn | Searches FASTA nucleotide queries in the input file against a nucleotide database |
| blastp | Searches FASTA protein queries in the input file against a protein database |
| blastx | Searches FASTA nucleotide queries in the input file, dynamically translated in all six frames, against a protein database and returns alignments at the protein level |
| blast_formatter | Formats a blast result using its assigned request ID (RID) or its saved archive file |
| convert2blastmask | Converts lowercase masking into makeblastdb readable data |
| deltablast | Searches a protein query against a protein database, using a more sensitive algorithm by taking conserved domain (delta cdd database required) matches into consideration |
| dustmasker | Masks the low complexity regions in the input nucleotide sequences |
| get_species_taxids.sh | Generates a list of species-level taxids (EntrezDirect installation required) for the input organism name or taxonomic ids above the species-level for use with a version 5 BLAST database in search limit or exclusion |
| legacy_blast.pl | Converts a legacy blast search command line into blast+ counterpart and execute it |
| makeblastdb | Formats input FASTA file(s) into a BLAST database |
| makembindex | Indexes an existing nucleotide database for use with indexed megablast search |
| makeprofiledb | Creates a conserved domain database from a list of input position specific scoring matrices (scoremats), usually generated by psiblast |
| psiblast | Finds members of a protein family, identifies proteins distantly related to the query, or builds position specific scoring matrix for the query through iterative rounds of searches |
| rpsblast | Searches a protein query against a conserved domain database to identify functional domains present in the query |
| rpstblastn | Searches a nucleotide query, by dynamically translated it in all six-frames first, against a conserved domain database |
| segmasker | Masks the low complexity regions in input protein sequences |
| tblastn | Searches a protein query against a nucleotide database dynamically translated in all six frames to return alignment at the protein level |
| tblastx | Searches a nucleotide query, dynamically translated in all six frames, against a nucleotide database translated in the same manner to return alignment at the protein level |
| update_blastdb.pl | Automatically downloads and decompresses all volumes of a specified preformatted blast databases from NCBI, AWS, or GCP (default is from NCBI) |
| windowmasker | Masks repeats found in input nucleotide sequences |
Test BLAST database
In addition to BLAST programs and accessory utilities, target database is an indispensable component of a standalone BLAST setup. The common set of pre-formatted NCBI BLAST databases is available as compressed archives from NCBI FTP site. Databases can also be prepared de novo from custom FASTA sequences locally using the makeblastdb utility. The best way to manage available BLAST databases is to place them in a dedicated directory. A subdirectory named "db" under the "C:\Users\taota\Desktop\blast-2.10.0+" directory is created for this, and its full path is "C:\Users\taota\Desktop\blast-2.10.0+\db".
Similar procedures in Figure 1 can be used to download preformatted BLAST databases from the NCBI ftp site to this dedicated database subdirectory. Her are the steps for downloading the single-volume 16S_ribosomal_RAN database databases:
- Point the browser to https://ftp.ncbi.nlm.nih.gov/blast/db/
- Right-click on the 16S_ribosomal_RNA.tar.gz file
- Select "Save target as …" from the popup menu (menus may differ among browsers)
- When prompted, use the "Save in" to change the directory to "C:\Users\taota\Desktop\blast-2.10.0+\db "
Use WinZip, 7-Zip, or other decompression utility to inflate the compressed archive first, then extract the files from the resulting archive. Note that the above steps download and install a database with a single volume. Large databases, such as nt, are provided as multi-volume sets. Get compressed archives with the same base name (with different ".##" or ".###" volume numbers) when attempting to reconstitute any multi-volume database. The database alias file, such as nt.nal or nr.pal, ties all volumes together back into the complete database. Also, for multi-volume databases, extra files enabling version 5 functionalities are only provided in the first volume. Figure 3 below shows an example inflation/extraction procedure using 7-Zip.
The blast+ package provides a Perl script, update_blastdb.pl, to help streamline the downloading of preformatted BLAST databases from NCBI. Downloading through this script requires the installation of the Perl package and execution from the command prompt under the database directory or "C:\users\taota\Desktop\blast-2.10.0+\db" in this example setup. The base command is:
perl update_blastdb.pl --passive –-decompress base_database_name
where "base_database_name" is the name of the target database without the "##.tar.gz" extension (e.g., refseq_rna, or refseq_representative_genomes).
Configuration
Further configuring the PC will help facilitate the execution of blast+ programs and streamline the access of installed databases. This configuration is through information stored in special user environment variables. For 2.10.0 release of the blast+ package, three variables are needed:
- A modified path environment variable to indicate the location of installed blast+ programs, with "C:\users\taota\Desktop\blast-2.10.0+\bin\;" prepended to its existing value
- A new BLASTDB environment variable as pointer to database location, with "C:\users\taota\Desktop\blast-2.10.0+\db\" as its value
- A new BLASTDB_LMDB_MAP_SIZE, with 1000000 as its value (needed to optimize makeblastdb operation when creating new database files)
Environment Variables
Here are the steps to create or modify environment variables to configure blast+ installation:
- Use toolbar’s search box to search for "edit user environment variables"
- Click the retrieved icon to launch the dialog box
- Click the "New…" button under the "User variable for ..." panel to create a new entry
- Type the variable name, then the absolute path (or other required input value)
- Click "OK" to close the prompts
- Highlight an existing variable, then click “Edit…” to edit its value
- Click "OK" to close the prompt and click "OK" again to exit
Example Screen Shots
Screen shots of these steps are shown, with the first two steps in Figure 4a, and the rest in Figure 4b.
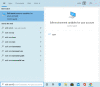
Figure 4a.
Configure standalone blast+ using Windows' environment variables: Use toolbar’s search box to find “Edit environment variable for your account,” then click to launch it.
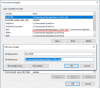
Figure 4b.
Configure standalone BLAST using Windows' environment variables: Use the “User variables for taota” section at the top of the dialog box to do the configuration. The two user variables relevant to blast+ are BLASTDB and path. Clicking (more...)
Execution and validation
Standalone blast+ programs do NOT have a graphical user interface (GUI) and must be executed from a command prompt window (CMD). The easiest way to open this prompt is to locate it by searching for “cmd” or “command prompt” using the toolbar’s search box as shown in Figures 5.
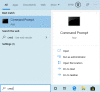
Figure 5.
Open a command prompt in Windows 7: Use the toolbar’s search box to search for the Command Prompt app. Click the icon to launch it.
Example Execution
Test the installation before using this installation for actual work. The test commands groups into three categories:
- Check blast-specific settings using commands
- Call blastdbcmd for database checking and specific sequence retrieval
- Call blastn to run a quick nucleotide blast search
- Get into the /db directory and use update_blastdb.pl to download NCBI databases
Detailed commands and their explanation
Technically, there are three components for a blast search, the input query, the target database, and the blast program. The test session below includes a set of representative commands, each testing a specific aspect of this example installation.
First section is on general checking and execution with marked command matching examples in Figure 6A.

Figure 6a.
The output of a work session testing the blast+ installation: The input commands are underlined with their function explained by inserted text.
Check the configuration by finding the settings of blast-specific environment variables (A).
set | find /I "blast"
The exact meaning of the command line is (from left to right) to:
- execute the set command (which dumps out all the environment variable settings)
- pass the console output ("|" pipe symbol) to next command
- execute find command in case-insensitive mode ("/I") to find lines containing blast in the output
Change the working directory to the installation (B) and check its content.
cd "C:\Users\taota\Desktop\blast-2.10.0+"
The exact meaning of the command line is to:
- a.
execute cd (change directory) command
- b.
set target the directory to that given in the argument
- c.
follow the above with the dir command (C, not shown) to check directory content
Check the manually downloaded 16S_ribosomal_RNA database with blastdbcmd (D):
blastdbcmd –db 16S_ribosomal_RNA –info
This command instructs the system to:
- execute blastdbcmd program
- look for a database call 16S_ribosomal_RNA (first in the BLASTDB specified directory, if failed, the current working directory)
- display the general information available for that database
Retrieve a sequence from this database for use as a test query (E):
blastdbcmd –db 16S_ribosomal_RNA -entry NR_025000 > test_16S_query.fa
This command instructs the system to:
- execute blastdbcmd program
- look for a database named 16S_ribosomal_RNA
- retrieve a sequence with its accession (-entry NR_025000) in default fasta format
- redirect (“>” symbol) the output to a text file name test_16S_query.fa
Check the version of the installation through the blastn program (F):
blastn -version
This command instructs the system to:
- execute blastn program
- display its version to the console
Run a nucleotide search using blastn and the test query sequence (G):
blastn –db 16S_ribosomal_RNA –query test_16S_query.fa -outfmt 7 -max_target_seqs 5
This command instructs the system to:
- execute blastn program
- search against the specified database 16S_ribosomal_RNA
- use sequence(s) in the specified file test_16S_query.fa as query
- ask for tabular ouput with header (-outfmt 7)
- request only the top 5 hits (-max_target_seqs 5)
- let the results display to the console (without specifying -out file_name)
The following focuses on database manipulation and example session is in Figure 6b.

Figure 6b.
The output of a work session focusing on database management and creation: The input commands are underlined with their function explained by inserted text.
Change working directory to the db subdirectory (H, not shown), then use update_blastdb.pl to download the preformatted swissprot database (I).
perl ..\bin\update_blastdb.pl --passive --decompress swissprot
This command instructs the system to:
- execute perl program (requires a separate installation)
- run the update_blastdb.pl script, which is in the parent directory’s bin subdirectory (..\bin\)
- use passive mode (--passive) for FTP
- decompress the downloaded file automatically (--decompress)
- set the requested database to swissprot
Check the extracted database files using dir (J, not shown), then download a multi-volume database refseq_rna using the same base command (K, not shown).
A common need to install standalone blast+ is to search against a custom collection of sequences. For that, the file with custom sequences in FASTA format needs to be converted to a blastable database using makeblastdb tool from the blast+ package.
Take advantage of the installed swissprot database to retrieve set of sequences from it and save them to a file (L) for use as example input.
blastdbcmd -db swissprot -taxids 9606 -out sp_hs_subset.faa
This command instructs the system to:
- execute blastdbcmd program
- set the source database to swissprot
- retrieves sequences based on their taxonomic id (-taxids 9606)
- save the output sequence to a file (-out sp_hs_subset.faa)
Call makeblastdb to convert the newly created FASTA sequence file into a blastable database (M).
makeblastdb -in sp_hs_subset.faa -dbtype prot -parse_seqids -title “demo: swissprot hs subset without taxid” -out sp_hs_subset
This command instructs the system to:
- execute makeblastdb program
- use sp_hs_subset.faa as input
- set the database type as protein
- index sequence ids (for specific sequence retrieval by id)
- create a title using text in the quotes
- rename the output (else the full FASTA file name including file extension will be used)
Check the resulted database using blastdbcmd (N, not shown).
Technical Assistance
Questions, feedback, and technical assistance requests should be sent to blast-help at:
vog.hin.mln.ibcn@pleh-tsalb
Questions on other NCBI resources should be addressed to NCBI Service Desk at:
vog.hin.mln.ibcn@ofni
- Contact the BLAST help desk: blast-help@ncbi.nlm.nih.gov
- Standalone BLAST Setup for Windows PC - BLAST® HelpStandalone BLAST Setup for Windows PC - BLAST® Help
Your browsing activity is empty.
Activity recording is turned off.
See more...