All Medical Genetics Summaries content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license which permits copying, distribution, and adaptation of the work, provided the original work is properly cited and any changes from the original work are properly indicated. Any altered, transformed, or adapted form of the work may only be distributed under the same or similar license to this one.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Pratt VM, Scott SA, Pirmohamed M, et al., editors. Medical Genetics Summaries [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012-.
Introduction
Simvastatin (brand name Zocor) is a member of the statin class of drugs, used to regulate low-density lipoprotein (LDL) cholesterol levels in various conditions, including familial hypercholesterolemia (FH), primary hyperlipidemia, hypertriglyceridemia, and primary dysbetalipoproteinemia. Statins are also used to reduce total mortality risk associated with coronary heart disease, non-fatal myocardial infarction, and revascularization procedures in adults at high risk of coronary heart disease events, including those with established vascular disease or diabetes. Approved by the US FDA for use primarily in adults, simvastatin is also approved for children aged 10 and older to manage FH (1). Administered as a pro-drug, simvastatin must be metabolized to simvastatin acid, which then acts in the liver and other tissues to reduce cholesterol production by competitively inhibiting HMG-CoA reductase (3-hydroxy-methylglutaryl-coenzyme). Simvastatin acid also promotes an increase in the uptake of LDL from the bloodstream, resulting in a reduction in cardiovascular risk and improved health outcomes for most individuals. However, individuals can experience adverse reactions; the most common are statin-associated musculoskeletal symptoms (SAMS) or other serious reactions.
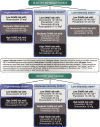
The Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG) of the Royal Dutch Association for the Advancement of Pharmacy indicate that individuals with decreased function in the organic anion transporting polypeptides 1B1 (OATP1B1) hepatic transport enzyme (encoded by the SLCO1B1 gene) have an increased risk of SAMS (2, 3). The CPIC guidelines provide dosing recommendations based on an individual’s predicted phenotype, stating that individuals with decreased or poor metabolizer phenotypes should be prescribed an alternative statin or a lower dose of simvastatin (Table 1) (2). Criteria for choosing the relative potency of an alternative statin or the dose of simvastatin are also outlined by CPIC (Figure 1) (2). The DPWG guidelines focus on the most common functional variant, a single nucleotide variation (SNV) at rs4149056, NM_006446.5:c.521T>C, recommending that individuals heterozygous or homozygous for the variant allele, resulting in decreased or poor function phenotype, choose an alternative statin (Table 2) (3, 4). The US FDA does not specifically address SLCO1B1 genetic variation in the simvastatin drug label, but it does discuss various medications that are either contraindicated (strong cytochrome P450 enzyme 3A4 [CYP3A4] inhibitors, gemfibrozil, cyclosporin and danazol) with simvastatin or may increase the risk of myopathy (1). The drug label for simvastatin in Switzerland, however, describes the increased risk of SAMS for individuals who have at least one variant allele at rs4149056, and recommends considering genotyping results indicating a CC genotype at this SNV during risk-benefit assessment before prescribing 80 mg doses of simvastatin due to higher myopathy risks (5). The interplay of genetics, co-medications, comorbidities, and simvastatin dose highlights the complex factors that contribute to an individual's risk of developing SAMS.
Table 1:
The Clinical Pharmacogenetics Implementation Consortium (CPIC) Recommendations for Simvastatin Based on SLCO1B1 Phenotype in Adults (2022)
| Phenotype | Implications | Dosing recommendation | Classification of recommendation | Considerations |
|---|---|---|---|---|
| SLCO1B1 increased function | Typical myopathy risk and statin exposure | Prescribe desired starting dose and adjust doses based on disease-specific guidelines. | Strong | The potential for drug–drug interactions and dose limits based on renal and hepatic function and ancestry should be evaluated before initiating a statin. |
| SLCO1B1 normal function | Typical myopathy risk and statin exposure | Prescribe desired starting dose and adjust doses based on disease-specific guidelines. | Strong | The potential for drug–drug interactions and dose limits based on renal and hepatic function and ancestry should be evaluated before initiating a statin. |
| SLCO1B1 decreased (or possible decreased) function | Increased simvastatin acid exposure as compared with normal function; increased risk of myopathy | Prescribe an alternative statin depending on the desired potencya. If simvastatin therapy is warranted, limit dose to <20 mg/day. | Strong | The potential for drug–drug interactions and dose limits based on renal and hepatic function should be evaluated before initiating a statin. The effects of drug–drug interactions may be more pronounced, resulting in a higher risk of myopathy. |
| SLCO1B1 poor function | Increased simvastatin acid exposure as compared with normal function; highly increased myopathy risk | Prescribe an alternative statin depending on the desired potencya. | Strong | The potential for
drug–drug Interactions and dose limits based on renal and hepatic function should be evaluated before initiating a statin. The effects of drug–drug interactions may be more pronounced, resulting in a higher risk of myopathy. |
Table 2:
The Dutch Pharmacogenetics Working Group (DPWG) Recommendations for Simvastatin Based on SLCO1B1 Genotype (2020)
| SLCO1B1 genotypea | Risk | Recommendation |
|---|---|---|
| 521CC | When using
simvastatin 80 mg/day, the risk of myopathy is increased 30-fold to
18% and the risk of severe myopathy is increased 48-fold to
12%. When using 40 mg/day, this risk is increased 7-fold to 1% and 11-fold to 0.68%. The gene variation leads to reduced simvastatin transport to the liver, which increases the simvastatin plasma concentration and therefore the risk of side effects. | Choose an
alternative. Consider any additional risk factors for statin-induced myopathy. Atorvastatin is affected less severely by the SLCO1B1 gene variation but is also affected by CYP3A4 inhibitors such as amiodarone, verapamil, and diltiazem. Use of atorvastatin is not recommended for individuals with additional risk factors for statin-induced myopathy. Rosuvastatin and pravastatin are influenced to a lesser extent by the SLCO1B1 gene variation. They are also not influenced by CYP3A4 inhibitors such as amiodarone, verapamil, and diltiazem. Fluvastatin is not significantly influenced by the SLCO1B1 gene variation or CYP3A4 inhibitors. |
| 521TC | When using
simvastatin 80 mg/day, the risk of myopathy is increased 5-fold to
3% for moderately severe to severe myopathy and 1.3% for severe
myopathy. When using 40 mg/day, this risk is increased 2.6-fold to 0.39% and 0.17%. The gene variation leads to reduced simvastatin transport to the liver, which increases the simvastatin plasma concentration and therefore the risk of side effects. | Choose an
alternative. Consider any additional risk factors for statin-induced myopathy. Atorvastatin is affected less severely by the SLCO1B1 gene variation but is also affected by CYP3A4 inhibitors such as amiodarone, verapamil, and diltiazem. Use of atorvastatin is not recommended for individuals with additional risk factors for statin-induced myopathy. Rosuvastatin and pravastatin are influenced to a lesser extent by the SLCO1B1 gene variation. They are also not influenced by CYP3A4 inhibitors such as amiodarone, verapamil, and diltiazem. Fluvastatin is not significantly influenced by the SLCO1B1 gene variation or CYP3A4 inhibitors. If an alternative is not an option: 1. Avoid simvastatin doses exceeding 40 mg/day (for example, by adding ezetimibe) 2. Advise the individual to report muscle symptoms. |
- a
DPWG literature review found association with only the T>C variation at rs4149056 (NM_006446.5:c.521T>C) and simvastatin side effects, though other gene variations were assessed (4).
This table adapted from (3) and personal communication from M. Nijenhuis (see Acknowledgements).
Drug: Simvastatin
Simvastatin, an HMG-CoA reductase inhibitor, is used with diet to manage FH, hypertriglyceridemia, primary dysbetalipoproteinemia, and to lower the risk of fatal cardiovascular events in individuals with coronary heart disease, cerebrovascular disease, peripheral vascular disease, diabetes, or a combination of these conditions. Simvastatin is primarily indicated for adults but is also approved for pediatric use in individuals aged 10 and older with FH. The recommended pediatric dose ranges from 10 mg to 40 mg daily, while adults are recommended 40 mg daily, with a rare exception of 80 mg daily only for those who have been taking this higher dose for more than 12 months without evidence of muscle toxicity. (1)
Statins are recommended as a first-line treatment for elevated LDL cholesterol levels or for diabetic individuals by the American College of Cardiology and American Heart Association (6) as well as by the American Association of Clinical Endocrinologists and American College of Endocrinology (7). Statins are contraindicated in individuals with acute liver failure or decompensated cirrhosis (1). Simvastatin should not be used with medications that strongly inhibit CYP3A4 (which includes macrolide antibiotics, some azole anti-fungal medications, antiviral medications, and nefazodone), nor with cyclosporine, danazol, or gemfibrozil (1). Consuming grapefruit juice while taking simvastatin can also increase the plasma levels of simvastatin due to inhibition of CYP3A4 (1, 5)
Statins, including simvastatin, work by inhibiting the HMG-CoA reductase enzyme, leading to decrease cholesterol production (8). This inhibition also reduces levels of mevalonate, which leads to upregulation of other enzymes involved in cholesterol biosynthesis, including the LDL receptor, further decreasing plasma LDL cholesterol levels (9). For individuals with FH due to LDL receptor loss of function, statins reduce cholesterol levels by decreasing the production of apolipoprotein-B containing lipoproteins in the liver (9). Decreased circulating lipid levels are associated with decreased risk of cardiovascular disease (CVD) and atherosclerotic CVD (ASCVD) (10). Simvastatin is absorbed by cells expressing the OATP1B1 drug transporter; in the liver, it is metabolized by CYP3A enzymes to simvastatin acid, which inhibits HMG-CoA reductase (11, 12). Simvastatin can be excreted from the liver via ABCB1 and ABCC2 transporters (11).
Higher doses of simvastatin and higher plasma levels of simvastatin are associated with higher rates of adverse effects, (12). including SAMS, a leading cause of drug discontinuation. The frequency of SAMS was found to directly correlate with simvastatin dose in clinical studies, ranging between 0.61–0.9% of individuals taking an 80 mg dose to 0.02–0.03% frequency with a 20 mg dose (1). Risk factors for SAMS include age of 65 or older, uncontrolled hypothyroidism, renal impairment, certain drug–drug interactions (discussed below), and Chinese ancestry (1). Individuals with reduced function in the drug-transporting enzyme OATP1B1 may be more likely to experience this toxicity (2, 3, 13) due to increased plasma concentration of simvastatin and its metabolites (14, 15). The CPIC guidelines stratify the relative risk of SAMS for statin use by decreased or poor OATP1B1 function (Figure 1) and are intended to be used with cardiovascular expert guidelines when selecting which statin and dose to use (see Therapeutic Recommendations Based on Genotype below for more information). The clinical presentation of SAMS can range from no muscle symptoms with serum creatine kinase (CK) elevations less than 4 times the upper normal limit (severity rating scale [SRM] 0) through the most severe presentation of immune-mediated necrotizing myositis (IMNM) (SRM 6). This standardized scale aids clinicians and researchers in understanding and managing SAMS (16).
Reports of muscle pain while taking statins may be a result of the nocebo effect, as suggested by a study of individuals with a history of SAMS. Out of 200 participants, 151 individuals reported similar frequencies of muscle pain during alternating periods of statin and placebo use, with two-thirds of the cohort resuming long-term statin use at the conclusion of the study (17). Balancing the concerns of SAMS with the therapeutic aim of statin therapy is important, as suboptimal statin use can increase cardiovascular event risk. Suboptimal use included discontinuation, nonadherence, dose reduction, and statin switching leading to reduced efficacy. One study in the UK reported that out of 1005 study participants, 156 individuals had suboptimal statin use and these individuals were at an increased risk of major adverse cardiovascular events (hazard ratio of 2.1) and all-cause mortality (hazard ratio 2.46) (18). Subgroup analysis determined that statin discontinuation or nonadherence were the major contributors to this risk (18).
Serious complications with simvastatin therapy include IMNM and hepatic dysfunction, and therapy should be discontinued if IMNM is suspected or serious hepatic injury (with clinical symptoms, jaundice, or both) occurs (1). Signs of IMNM include proximal muscle weakness, elevated serum CK persisting after statin discontinuation, and necrotizing myopathy on muscle biopsy (1). Two myositis-specific autoantibodies can also be detected during IMNM: anti-signal recognition particle and anti-HMG-CoA Reductase antibodies (19, 20). Individuals with IMNM may require treatment with immunosuppressive agents (1). More common but less severe side effects include upper respiratory infection, headache, abdominal pain, constipation, and nausea (1). There have also been reports of elevated hemoglobin A1c (HbA1C) and fasting glucose levels with statin therapy (1). Simvastatin is not recommended during pregnancy due to decreased synthesis of cholesterol and other biologically active substances derived from cholesterol (1). The specific therapeutic needs of the individual should be considered, though the FDA-approved label states that hyperlipidemia treatment during pregnancy is generally not necessary, given the chronic nature of the atherosclerotic process (1). Clinical information does not indicate a drug-associated risk of major congenital malformations if used during pregnancy, but there is insufficient evidence to evaluate the risk of miscarriage associated with simvastatin therapy (1). There is also a lack of data on the safety of simvastatin use during breastfeeding, and the consensus for clinical practice is to avoid its use by breastfeeding mothers (21). Some advocate for the continued use of statins by expectant mothers and women attempting to conceive with FH due to the increase in lipid levels during pre-conception and pregnancy (12–15 months per pregnancy) (22). Additionally, adverse pregnancy outcomes (including gestational hypertension or diabetes, preeclampsia, or pre-term birth) can increase ASCVD risk, and the natural lipid fluctuations during pregnancy may put individuals with elevated baseline cholesterol levels at greater risk (23). More clinical research is needed to assess the risk of various statins with adverse maternal and fetal or neonatal outcomes (23).
The use of statins in pediatric individuals has been shown to be safe and effective for individuals with heterozygous FH aged 10 and older. A controlled clinical study showed no significant effect on growth or sexual maturation in male (n=99) or female (n=76, all one-year post-menarche) participants (1). Additional studies have examined the efficacy and side effects of varying dosing of simvastatin in pediatric individuals with FH, and reported no clinically relevant side effects (24). Statin therapy in a pediatric population appears safe, though continuity of care remains important as these individuals transition from pediatric to adult clinical care (25).
Individuals aged 65 and over are at increased risk of adverse effects of simvastatin (1). Pharmacokinetic studies suggest that individuals aged 70–78 have a 45% higher plasma level of simvastatin compared with those aged 18–30. Individuals aged 65 and older taking 80 mg simvastatin are at increased risk of myopathy compared with those under 65 years of age taking the same high dose (1). A separate retrospective study reported that high-dose statin use (including simvastatin) in individuals aged 65 and older was associated with an increased burden of myalgia and elevated liver enzymes compared with a control cohort taking low-dose statins (26). Hepatic and renal impairment, often observed in geriatric individuals, are also risk factors for SAMS regardless of age (1).
Gene: SLCO1B1
The SLCO1B1 gene, located on the short arm of chromosome 12, encodes OATP1B1, one of the major hepatic influx transport proteins. Primarily expressed on the basolateral surface of hepatocytes, OATP1B1 facilitates the uptake of various endogenous and exogenous compounds including bile acids, thyroid hormones, bilirubin, methotrexate, angiotensin-converting enzyme inhibitors, angiotensin II receptor blockers, and statins. Decreased expression or function of OATP1B1 can lead to increased plasma concentrations of these substrates, potentially resulting in systemic drug toxicity or adverse effects. Genetic variations at the SLCO1B1 locus, as well as drug–drug interactions, can change OATP1B1 function and increase an individual’s risk for adverse effects from these substrate medications. (27, 28)
The Pharmacogene Variation Consortium (PharmVar) maintains a standardized nomenclature of SLCO1B1 variants, commonly called star (*) alleles (27). Each star allele is defined by a core variant or variants inherited together on a single allele, or haplotype, with the pair of alleles present in any individual called their diplotype or genotype. Star alleles are assigned a clinical function level by CPIC. Of the 44 star alleles defined by PharmVar, only 13 SLCO1B1 star alleles have been assigned a clinical functional status by CPIC (Table 3) (29, 30). The SLCO1B1*1 and *37 alleles are classified as normal function, while multiple alleles such as SLCO1B1*5, *9, and *15, are classified as having no function. The SLCO1B1*14 and *20 alleles are classified as increased-function alleles (29). It is common in pharmacogene nomenclature to assign the *1 allele as the reference allele and the baseline for “normal” allele function. Also, *1 is the presumed allele when no variants are identified by testing.
Table 3:
Functional Classification of SLCO1B1 Allele (CPIC, 2021)
| Allele clinical functional status | SLCO1B1 star alleles |
|---|---|
| Increased function | *14, *20 |
| Normal function | *1, *37 |
| No function | *5, *9, *15, *23, *31, *46, *47, *48, *49 |
| Unknown function | *43, *44, *45 |
Table adapted from (29). CPIC - Clinical Pharmacogenetics Implementation Consortium
The normal-function SLCO1B1*37 allele is defined by a SNV that substitutes an aspartic acid (D) for asparagine (N) at position 130 of the protein (rs2306283, NM_006446.5:c.388A>G) with no other functionally significant variants (29, 31). This variant is present in 18 other star alleles with varying clinical functions, underscoring the importance of testing the breadth of known star alleles for accurate genotyping (27). Another commonly studied SNV is rs4149056 (NM_006446.5:c.521T>C), found in the SLCO1B1*5, SLCO1B1*15, SLCO1B1*40, SLCO1B1*46, and SLCO1B1*47 alleles. All but one of these alleles are classified as no function by CPIC—the one outlier allele, SLCO1B1*40, has uncertain function due to the presence of other variants with unknown functional impact (27, 30). The rs4149056 variant is found in 4 out of the 9 no-function alleles. The remaining 5 no-function alleles are defined by other SNVs or structural variants resulting in either a full or partial gene deletion (31).
Historic star allele designations have been reviewed and standardized by PharmVar, leading to some alleles being reclassified as sub-alleles based on shared core variants and biochemical functional status (27). The biochemical activity of a protein does not automatically translate to the same level of clinical activity; thus, in vitro pharmacokinetic/pharmacodynamic studies identifying hypo-active forms of OATP1B1 may ultimately result in a clinical phenotype no different than a complete loss-of-function allele (27).
An individual’s SLCO1B1 genotype (pair of inherited alleles) can predict their phenotype (Table 4) (32). Individuals with normal or increased function OATP1B1 transporter phenotype are at a lower risk of adverse drug reactions, while those with poor function or decreased function phenotypes are at higher risk of adverse reactions due to higher plasma concentrations of the substrate medication and its metabolites. An individual is assigned a phenotype of “possible decreased function” when one known no-function allele is present, and the second allele has unknown or uncertain function (2).
Table 4:
Selected SLCO1B1 Phenotype-Genotype Predictions (CPIC, 2022)
| Clinical phenotype | Example SLCO1B1 genotype | Clinical alert categorya |
|---|---|---|
| Normal function | *1/*1 *1/*14 *1/*37 | Low risk |
| Increased function | *14/*14 *14/*20 *20/*20 | Low risk |
| Decreased function | *1/*5 *1/*9 *15/*37 | High risk |
| Possible decreased function | *2/*5 *2/*15 *3/*9 | High risk |
| Poor function | *5/*5 *5/*9 *9/*23 *15/*15 | High risk |
- a
These genotypes and phenotypes are recommended by CPIC to be flagged as low or high risk when implemented in electronic health records notifications. CPIC - Clinical Pharmacogenetics Implementation Consortium
This table is adapted from (32).
The frequency of SLCO1B1 alleles can vary significantly based on population-specific genetic ancestry. However, allele frequencies cited reported in the literature may not fully represent allelic variation within a population. Owing to the recent reclassification of some historic star alleles and the underrepresentation of many populations in genetic and genomic research, these frequencies should be considered cautiously (27). For example, studies of populations living in the United Arab Emirates or Qatar examined only one or 2 SNVs and assigned the most common genotype based on those variants, excluding other star alleles (33, 34). Additional studies using current PharmVar allele definitions are needed across multiple populations to understand allele and phenotype frequencies for SLCO1B1. The following information should be interpreted as approximate frequencies based on existing publications.
The SLCO1B1*1 allele is clinically determined to be a normal-function allele, with a frequency of 50% or less for all CPIC biogeographical groups (35). However, the SLCO1B1*37 allele—previously called *1B and also a normal-function allele—is the most frequently reported haplotype in African-American and Sub-Saharan-African populations, with a frequency of 76–80%, and 60% in East Asian populations (35).
Reduced- and increased-function alleles are less common globally. The no-function allele SLCO1B1*15 is more common in many biogeographical groups compared with SLCO1B1*5. The combined frequency of both can range from 24% in American, 20% in Near Eastern, 17% in European, 12% in East Asian, 7% in Central/South Asian, 1–2% in African-American/Afro-Caribbean and Sub-Saharan populations, to not observed in Oceanian populations (35). The SNV that is shared between SLCO1B1*5 and SLCO1B1*15 was observed at a frequency of 24% in a Qatari population of mixed genetic ancestry, while the SNV associated with SLCO1B1*37 was found in approximately 50% of study participants (33). Other no-function alleles are observed far less frequently, such as SLCO1B1*9 due to variation at rs59502379, which has an allele frequency of 0–4.6% of the global populations in the Allele Frequency Aggregate (ALFA) data (36). The increased-function SLCO1B1*14 allele has been reported almost exclusively in European genetic backgrounds at a frequency of 12% (35).
Variations in SLCO1B1 causes altered transportation of endogenous substances such as bilirubin. The rare, inherited disorder Rotor syndrome is caused by bi-allelic loss of function variants in both SLCO1B1 and SLCO1B3, which encodes another organic anion transporting polypeptide (37). Rotor syndrome presents with a benign form of hyperbilirubinemia and jaundice, clinically indistinguishable from Dubin-Johnson syndrome, though the elevated bilirubin seen in Rotor syndrome is a mix of both conjugated and unconjugated forms (38). Inherited in an autosomal recessive pattern, Rotor syndrome does not require therapy, but diagnosis is important to ensure other, more serious hepatobiliary disorders are not the cause of the jaundice (38).
Phenoconversion
Phenoconversion occurs when certain medications inhibit OATP1B1 activity, reducing transport of various substrates into hepatocytes and leading to a lower activity phenotype than predicted by genotype alone. Several drugs have been identified by in vitro assays to inhibit OATP1B1 including atorvastatin, cyclosporin, digoxin, gemfibrozil, ketoconazole, and rifampin among others (28). In vivo administration of cyclosporine with rosuvastatin or fluvastatin resulted in increased statin exposure, presumably due to decreased function of OATP and CYP3A4 (39, 40, 41). The US FDA’s approved label for simvastatin states that concomitant use of simvastatin with cyclosporine, danazol, or gemfibrozil is contraindicated due to increased risk of myopathy (1).
Wojtyniak and colleagues have made available their drug–drug gene interaction data in the form of a clinical decision support tool at https://nemos.shinyapps.io/simvastatin_simulator/. However, the website states that “decisions on therapeutics and dosing recommendations should not exclusively be build[sic] based on the results of the simvastatin exposure simulator and do not replace clinical judgement” (42).
Linking SLCO1B1 Genetic Variation with Treatment Response
The risk of SAMS for simvastatin correlates with SLCO1B1 variants that reduce OATP1B1 function, thereby increasing an individual’s exposure to simvastatin and its metabolites. Increased and normal function SLCO1B1 haplotypes do not have a clinically significant impact on simvastatin metabolism or increase the risk of adverse effects above an individual’s baseline risk (2). The variant at rs4149056 (c.521T>C; the defining SNV for SLCO1B1*5 and also a part of SLCO1B1*15) was found to increase simvastatin acid exposure (as measured by total plasma concentration over time, or area under the curve) by approximately 40% in healthy volunteers, increasing the risk of SAMS (14). This variant also increased exposure to simvastatin acid in a pediatric population (43), though the CPIC found insufficient data to make pediatric-specific recommendations (2). A pooled analysis of 11 studies on the association of rs4149056 variation and SAMS risk in Caucasians with the CC or TC genotype indicated a higher risk of myopathy, with the CC genotype carriers having a 2.81 odds ratio (OR) for myopathy compared with the wildtype (TT) genotype, and heterozygotes (CT genotype) having an OR of 1.78 (13). The pooled analysis of studies in Asian populations, though including only 2 publications, reported a 1.8 times higher risk of SAMS when the C allele was present (13). Increased plasma levels of simvastatin acid were also reported to be associated with SLCO1B1 variants at rs11045819, (c.463C>A, which is the defining SNV for SLCO1B1*4) and rs34671512 (c.1929A>C, one of 2 SNVs in SLCO1B1*20) (12).
O’Brien and colleagues, in a retrospective analysis of more than 11,000 medical records over 5 years, found that, after correcting for covariates, individuals who self-reported as “Black/African-American” or “other/multiple race” were less likely to experience adverse reactions to simvastatin (44). This study also found that lower age and comorbid hypertension were significant covariates for increased probability of adverse reaction to simvastatin. However, the authors noted a relatively small number of adverse events and acknowledged the limitations off often incomplete electronic health record data (44). A study of nearly 300 individuals with FH found that variation at rs4149056 (c.521T>C) was not associated with SAMS; instead, increased age was the most significant risk factor (45). The US FDA’s approved label also advises that advanced age (65 years and older) is a risk factor for SAMS (1).
Increased risk of developing type-2 diabetes is another side effect of statin use (46, 47, 48), and variation at rs4149056 has been associated with an increased risk, as indicated by elevated hemoglobin A1C levels (34). However, a large study of over 7,500 individuals (1,373 individuals treated with statins and 6,415 not treated with statins) found no association between rs4149056 T>C variation and incidence of diabetes or changes in blood glucose levels (15).
Studies examining the link between SLCO1B1 genetic variation and the efficacy of statins reported that decreased OATP1B1 transport (most often studied in the context of rs4149056, c.521T>C genotype) correlates with an attenuated effect on the lipid-lowering capability of statins, including simvastatin. However, the effect is small (<10 mg/dl) and unlikely to impact the frequency of vascular events, leading CPIC to base their recommendations primarily on the risk of SAMS and pharmacokinetic data (see Supplement of (2)) rather than on efficacy concerns. Many studies reported a milder reduction in cholesterol in individuals with one or 2 copies of the variant rs4149056 c.521T>C allele, though it is unclear if this is due to medication nonadherence (specifically due to myopathy or other adverse effects), differences in genetic background, reduced OATP1B1 function, or a combination of these factors (49, 50, 51, 52, 53, 54, 55).
In contrast, no association was found between SLCO1B1 variants at rs4149056 (c.521T>C), rs2306283 (c.388A>G), or rs4363657 (g.89595T>C) and lipid levels following simvastatin therapy in a cohort of nearly 400 Thai individuals with hypercholesterolemia (56). Similarly, a review of several studies in Brazil found no association between SLCO1B1 variants (namely, the SLCO1B1*5, *15, *4, and *14 alleles) and efficacy of atorvastatin or simvastatin (57).
Additional Genes of Interest
Given the role of CYP3A enzymes in metabolizing simvastatin to simvastatin acid, it is not surprising that some studies have examined the association between CYP3A4 variation and simvastatin pharmacokinetics. Like other cytochrome P450 family members, genetic variation at the CYP3A loci can result in decreased or no function of the encoded enzyme. While CPIC has not assigned a clinical function to CYP3A4 haplotypes (58), the biochemical function has been used to predict metabolism phenotypes of intermediate or poor CYP3A4 metabolizers (12). One study reported a genome wide association with CYP3A4*2 (rs55785340, c.664T>C) genotype, assigned as an intermediate metabolizer phenotype, and increased simvastatin acid exposure (12). Individuals heterozygous for the CYP3A4*22 or CYP3A5*3 alleles (classified as intermediate metabolizers) had significantly higher plasma simvastatin concentrations and lower total and plasma LDL cholesterol levels (59). However, Kitzmiller and colleagues reported no association between CYP3A4*22 (a decreased-function allele) or CYP3A5*3 (clinically categorized by CPIC as a no-function allele (60)) and the cholesterol-lowering response to simvastatin (50).
Further research found a connection between statin response and variations in the hepatic efflux transporter ABCB1 and a leukocyte immunoglobin receptor locus, LILRB5 (61). The SNV at rs1045642 in the ABCB1 gene, resulting in a synonymous protein change, was linked to a significant reduction in non-high-density lipoprotein (HDL) levels in a recessive inheritance model. The LILRB5 variant rs12975366 showed effects in a dominant fashion, leading to a more pronounced decrease in non-HDL levels. These 2 loci appear to have a synergistic effect, although further studies are needed for confirmation.
An analysis of the UK Biobank data indicated a potential association between NAT2 genetic variation and statin use. The NAT2 locus encodes N-acetyltransferase 2 and has been liked to abnormal sensitivity to amifampridine. Wendt and colleagues reported an association between the NAT2*5 allele and LDL cholesterol levels, with diplotypes including this allele being more common among statin users than non-users. They proposed a model where the NAT2 enzyme might acetylate LDL cholesterol, affecting its binding to LDL receptors and indirectly impacting statin efficacy (62).
An association has been observed between variants in HLA loci and IMNM in a Japanese cohort. Several variant alleles were found to be present at a higher rate in individuals with IMNM, although the study did not specifically assess statins as a trigger for IMNM. Candidate HLA-risk alleles for IMNM include A*02:07, B*46, C*01:02, DRB1*08:03, DRB1*11:01, DQB1*06:01, DPB1*02:02, DPB1*05:01. (20)
Genetic Testing
The NIH Genetic Testing Registry (GTR) offers tests for simvastatin response and SLCO1B1 genetic variation. Considering the recent reclassification of SLCO1B1 alleles by PharmVar, it is important to consider the specific testing methodology and review the genotype-phenotype assignments. Targeted SNV genotyping may only examine the most common functional variants without distinguishing between known haplotypes of differing functional status, such as SLCO1B1*5 versus SLCO1B1*40. Gene sequencing may not detect structural variations or changes in copy number. Resources such as the Genotype Selection Interface (GSI) from PharmGKB, the Pharmacogenomics Clinical Annotation Tool (PharmCAT), and the PharmVar SLCO1B1 allele definitions assist in report interpretation. The decision to test for SLCO1B1 variation before or during standard -dose simvastatin therapy depends on the managing clinician. The FDA has no requirement to assay SLCO1B1 before therapy nor does CPIC issue guidance regarding testing. However, the DPWG recommends genotyping before starting an 80 mg/day dose of simvastatin for drug tolerance, and considers testing beneficial at a 40 mg/day dose, advising testing before or directly after initiating treatment (4). The data from PharmGKB for the Swissmedic drug labeling recommends SLCO1B1 genotype testing with simvastatin therapy, both alone and with ezetimibe (11).
The clinical validity of SLCO1B1 genotyping in predicting SAMS has been assessed. The SEARCH study, which focused on individuals with a history of myocardial infarction taking 80 mg simvastatin, found that individuals with at least one C allele at rs4149056, had a significantly increased risk of myopathy over 5 years, with an OR of 4.5 per C allele (63). An analysis of this study showed that at least one C allele found by genotyping has a positive predictive value (PPV) for myopathy risk of 4.1%, a negative predictive value (NPV) of 99.4%, a specificity of 73.7% and sensitivity of 70.4%; biallelic C genotype at this SNV has a PPV of 18.6%, NPV of 99.8%, specificity of 98.3%, and sensitivity of 25.1% (64). No additional risk loci for SAMS have been identified (65).
One study reported that utilization of SLCO1B1 pharmacogenetic testing within a clinical decision support tool led to fewer prescriptions of medications that increase the risks of SAMS for individuals with SLCO1B1 at-risk genotypes. This information is more likely to result in the cancellation of a simvastatin prescription if the genotype is known before prescribing rather than afterward (66). A similar study found that clinical testing and reporting of the SLCO1B1 genotype to guide simvastatin therapy resulted in noninferior outcomes for the genotype-guided group, with no instances of simvastatin prescribing for known OATP1B1 decreased or poor function phenotypes, suggesting that, given the information in advance, physicians avoid prescribing simvastatin to individuals with an at-risk genotype (67).
The SLCO1B1 Gene Interactions with Medications Used for Additional Indications
The FDA has included the SLCO1B1 gene on very few drug labels to date; including rosuvastatin (another statin), elagolix, and viloxazine (68). Elagolix is a gonadotropin-releasing hormone antagonist medication used to manage pain associated with endometriosis (69). Viloxazine is a selective norepinephrine reuptake inhibitor used for attention deficit hyperactivity disorder (70). Guidelines are available from CPIC on SLCO1B1 and multiple statins,(2) and PharmGKB, CPIC, and the FDA also provide additional information on gene-drug interactions involving SLCO1B1 (search for “SLCO1B1”).
Therapeutic Recommendations based on Genotype
This section contains excerpted1 information on gene-based dosing recommendations. Neither this section nor other parts of this review contain the complete recommendations from the sources.
2023 Statement from the US Food and Drug Administration (FDA):
Warnings and Precautions- Myopathy and Rhabdomyolysis
Simvastatin may cause myopathy and rhabdomyolysis… Risk factors for myopathy include age 65 years or greater, uncontrolled hypothyroidism, renal impairment, concomitant use with certain other drugs (including other lipid lowering therapies), and higher simvastatin dosage; Chinese patients on simvastatin may be at higher risk for myopathy…The risk of myopathy is increased by elevated plasma levels of simvastatin and simvastatin acid. The risk is also greater in patients taking an 80 mg daily dosage of simvastatin compared with patients taking lower simvastatin tablets dosages and compared with patients using other statins with similar or greater LDL-C lowering efficacy.
Steps to Prevent or Reduce the Risk of Myopathy and Rhabdomyolysis
The concomitant use of strong CYP3A4 inhibitors with simvastatin is contraindicated. If short-term treatment with strong CYP3A4 inhibitors is required, temporarily suspend simvastatin during the duration of strong CYP3A4 inhibitor treatment. The concomitant use of simvastatin with gemfibrozil, cyclosporine, or danazol is also contraindicated … Simvastatin dosage modifications are recommended for patients taking lomitapide, verapamil, diltiazem, dronedarone, amiodarone, amlodipine or ranolazine.
Please review the complete therapeutic recommendations that are located here: (1)
2022 Statement from the Clinical Pharmacogenetics Implementation Consortium (CPIC):
Phenotype: SLCO1B1 decreased function or SLCO1B1 possible decreased function
Implications: Increased simvastatin acid exposure as compared with normal function; increased risk of myopathy.
Dosing recommendation: Prescribe an alternative statin depending on the desired potency (see Figure 1 for recommendations for alternative statins). If simvastatin therapy is warranted, limit dose to <20 mg/day.
Phenotype: SLCO1B1 poor function
Implications: Increased simvastatin acid exposure compared with normal and decreased function; highly increased myopathy risk.
Dosing recommendation: Prescribe an alternative statin depending on the desired potency (see Figure 1 for recommendations for alternative statins)
Please review the complete therapeutic recommendations that are located here: (2)
2020 Summary of recommendations from the Dutch Pharmacogenetics Working Group (DPWG) of the Royal Dutch Association for the Advancement of Pharmacy (KNMP)
SLCO1B1 521CC: [simvastatin]
When using simvastatin 80 mg/day, the risk of myopathy is increased 30-fold to 18% and the risk of severe myopathy is increased 48-fold to 12%. When using 40 mg/day, this risk is increased 7-fold to 1% and 11-fold to 0.68% respectively. The gene variation leads to reduced simvastatin transport to the liver, which increases the simvastatin plasma concentration and therefore the risk of side effects.
1. Choose an alternative
Consider any additional risk factors for statin-induced myopathy.
Atorvastatin is affected less severely by the SLCO1B1 gene variation, but is also affected by CYP3A4 inhibitors such as amiodarone, verapamil and diltiazem. Use of atorvastatin is not recommended for patients with additional risk factors for statin-induced myopathy.
Rosuvastatin and pravastatin are influenced to a lesser extent by the SLCO1B1 gene variation. They are also not influenced by CYP3A4 inhibitors such as amiodarone, verapamil and diltiazem.
Fluvastatin is not significantly influenced by the SLCO1B1 gene variation or CYP3A4 inhibitors.
SLCO1B1 521TC: [simvastatin]
When using simvastatin 80 mg/day, the risk of myopathy is increased 5-fold to 3% for moderately severe to severe myopathy and 1.3% for severe myopathy. When using 40 mg/day, this risk is increased 2.6-fold to 0.39% and 0.17% respectively. The gene variation may lead to reduced simvastatin transport to the liver, which may increase simvastatin plasma concentrations and therefore the risk of side effects.
1. Choose an alternative
Consider any additional risk factors for statin-induced myopathy.
Atorvastatin is affected less severely by the SLCO1B1 gene variation, but is also affected by CYP3A4 inhibitors such as amiodarone, verapamil and diltiazem. Use of atorvastatin is not recommended for patients with additional risk factors for statin-induced myopathy.
Rosuvastatin and pravastatin are influenced to a lesser extent by the SLCO1B1 gene variation. They are also not influenced by CYP3A4 inhibitors such as amiodarone, verapamil and diltiazem.
Fluvastatin is not significantly influenced by the SLCO1B1 gene variation or CYP3A4 inhibitors.
2. If an alternative is not an option:
1. Avoid simvastatin doses exceeding 40 mg/day (for example, by adding ezetimibe)a
2. Advise the patient to report muscle symptoms. a
Please review the complete therapeutic recommendations that are located here: (3) a Note that minor variations in wording versus the cited guidelines are included here based on personal communication from DPWG.
Nomenclature for Selected SLCO1B1 Alleles
| Common allele name | Alternative names | HGVS reference sequence | dbSNP reference identifier for allele location | |
|---|---|---|---|---|
| Coding | Protein | |||
| SLCO1B1*1 | SLCO1B1*1A | NM_006446.5 | NP_006437.3 | |
| SLCO1B1*4 | 35305C>A (P155T) | NM_006446.5:c.463C>A | NP_006437.3:p.Pro155Thr | rs11045819 |
| SLCO1B1*5 | 37041T>C (V174A) | NM_006446.5:c.521T>C | NP_006437.3:p.Val174Ala | rs4149056 |
| SLCO1B1*9 | 64425G>C (G488A) | NM_006446.5:c.1463G>C | NP_006437.3:p.Gly488Ala | rs59502379 |
| SLCO1B1*14a | 35230A>G (N130D) | NM_006446.5:c.388A>G | NP_006437.3:p.Asn130Asp | rs2306283 |
| 35305C>A (P155T) | NM_006446.5:c.463C>A | NP_006437.3:p.Pro155Thr | rs11045819 | |
| SLCO1B1*15b | 35230A>G (N130D) | NM_006446.5:c.388A>G | NP_006437.3:p.Asn130Asp | rs2306283 |
| 37041T>C (V174A) | NM_006446.5:c.521T>C | NP_006437.3:p.Val174Ala | rs4149056 | |
| SLCO1B1*20c | 35230A>G (N130D) | NM_006446.5:c.388A>G | NP_006437.3:p.Asn130Asp | rs2306283 |
| 97468A>C (L643F) | NM_006446.5:c.1929A>C | NP_006437.3:p.Leu643Phe | rs34671512 | |
| SLCO1B1*37 | SLCO1B1-*1B,
*1F, *1G, *1H 35230A>G (N130D) | NM_006446.5:c.388A>G | NP_006437.3:p.Asn130Asp | rs2306283 |
- a
SLCO1B1*14 represents a consolidated core haplotype encompassing the previously named SLCO1B1*14 and SLCO1B1*18 alleles.
- b
SLCO1B1*15 represents a consolidated core haplotype encompassing the previously named SLCO1B1*15A, SLCO1B1*15B, and SLCO1B1*17 alleles.
- c
SLCO1B1*20 represents a consolidated core haplotype encompassing the previously named SLCO1B1*20, SLCO1B1*21, and SLCO1B1*35 alleles.
Pharmacogenetic Allele Nomenclature: International Workgroup Recommendations for Test Result Reporting (71).
Guidelines for the description and nomenclature of gene variations are available from the Human Genome Variation Society (HGVS).
Acknowledgments
The author would like to acknowledge Michael Asger Andersen, MD, PhD, Department of Clinical Pharmacology, Copenhagen University Hospital - Bispebjerg and Frederiksberg, Copenhagen, Denmark; Marga Nijenhuis, PhD, Royal Dutch Pharmacists Association (KNMP), The Hague, The Netherlands; and Jeffrey A. Shaman, PhD, MS, Chief Science Officer, Coriell Life Sciences, Philadelphia, PA, USA for providing an expert review of this summary.
References
- 1.
- SIMVASTATIN- simvastatin tablet. Somerset, NJ, USA: Micro Labs Limited; 2023. 'Available from:' https://dailymed
.nlm .nih.gov/dailymed/drugInfo .cfm?setid=5c1c694c-4b08-469e-b538-08e69df06146. - 2.
- Cooper-DeHoff, R.M., et al., The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin Pharmacol Ther, 2022. 111(5): p. 1007-1021. [PMC free article: PMC9035072] [PubMed: 35152405]
- 3.
- Pharmacogenetic Recommendation Text [Cited 11 Dec 2023]. Available from https://www
.knmp.nl/dossiers /farmacogenetica. - 4.
- SLCO1B1: simvastatin [Cited 11 Dec 2023]. Available from https://www
.g-standaard .nl/risicoanalyse/M0003981.pdf. - 5.
- Organon GmbH. Zocor (R) 2023 June 2023 16 Feb 2024]; Available from: https://amiko
.oddb.org/de/fi?gtin=49742. - 6.
- Correction to: 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation, 2019. 140(11): p. e649-e650. [PubMed: 31498691]
- 7.
- Jellinger, P.S., et al., American Association of Clinical Endocrinologists and American College of Endocrinology Guidelines for Management of Dyslipidemia and Prevention of Cardiovascular Disease. Endocr Pract, 2017. 23(Suppl 2): p. 1-87. [PubMed: 28437620]
- 8.
- Talreja, O., C.C. Kerndt, and M. Cassagnol, Simvastatin, in StatPearls. 2024: Treasure Island (FL). Available from https://www
.ncbi.nlm .nih.gov/pubmed/30422514. [PubMed: 30422514] - 9.
- Pedersen, T.R. and J.A. Tobert, Simvastatin: a review. Expert Opin Pharmacother, 2004. 5(12): p. 2583-96. [PubMed: 15571475]
- 10.
- Soppert, J., et al., Lipoproteins and lipids in cardiovascular disease: from mechanistic insights to therapeutic targeting. Adv Drug Deliv Rev, 2020. 159: p. 4-33. [PubMed: 32730849]
- 11.
- PharmGKB. SLCO1B1 Drug Label Annotations. 2023 13 Dec 2023]; Available from: https://www
.pharmgkb .org/gene/PA134865839/labelAnnotation. - 12.
- Mykkanen, A.J.H., et al., Genomewide Association Study of Simvastatin Pharmacokinetics. Clin Pharmacol Ther, 2022. 112(3): p. 676-686. [PMC free article: PMC9540481] [PubMed: 35652242]
- 13.
- Turongkaravee, S., et al., A systematic review and meta-analysis of genotype-based and individualized data analysis of SLCO1B1 gene and statin-induced myopathy. Pharmacogenomics J, 2021. 21(3): p. 296-307. [PMC free article: PMC8159730] [PubMed: 33608664]
- 14.
- Jiang, F., et al., The influences of SLCO1B1 and ABCB1 genotypes on the pharmacokinetics of simvastatin, in relation to CYP3A4 inhibition. Pharmacogenomics, 2017. 18(5): p. 459-469. [PubMed: 28350522]
- 15.
- Fernandes Silva, L., et al., Effects of SLCO1B1 Genetic Variant on Metabolite Profile in Participants on Simvastatin Treatment. Metabolites, 2022. 12(12). [PMC free article: PMC9785662] [PubMed: 36557197]
- 16.
- Alfirevic, A., et al., Phenotype standardization for statin-induced myotoxicity. Clin Pharmacol Ther, 2014. 96(4): p. 470-6. [PMC free article: PMC4172546] [PubMed: 24897241]
- 17.
- Herrett, E., et al., Statin treatment and muscle symptoms: series of randomised, placebo controlled n-of-1 trials. BMJ, 2021. 372: p. n135. [PMC free article: PMC7903384] [PubMed: 33627334]
- 18.
- Turner, R.M., et al., Investigating the prevalence, predictors, and prognosis of suboptimal statin use early after a non-ST elevation acute coronary syndrome. J Clin Lipidol, 2017. 11(1): p. 204-214. [PMC free article: PMC5399750] [PubMed: 28391887]
- 19.
- Ma, X. and B.T. Bu, Anti-SRP immune-mediated necrotizing myopathy: A critical review of current concepts. Front Immunol, 2022. 13: p. 1019972. [PMC free article: PMC9612835] [PubMed: 36311711]
- 20.
- Ohnuki, Y., et al., Association of immune-mediated necrotizing myopathy with HLA polymorphisms. HLA, 2023. 101(5): p. 449-457. [PubMed: 36565042]
- 21.
- Simvastatin, in Drugs and Lactation Database (LactMed(R)). 2006: Bethesda (MD). Available from https://www
.ncbi.nlm .nih.gov/pubmed/30000419. - 22.
- Holmsen, S.T., et al., Statins and breastfeeding in familial hypercholesterolaemia. Tidsskr Nor Laegeforen, 2017. 137(10): p. 686-687. [PubMed: 28551957]
- 23.
- Grant, J.K., et al., Lipid-Lowering Therapy in Women of Childbearing Age: a Review and Stepwise Clinical Approach. Curr Cardiol Rep, 2022. 24(10): p. 1373-1385. [PubMed: 35904667]
- 24.
- Dirisamer, A., et al., The effect of low-dose simvastatin in children with familial hypercholesterolaemia: a 1-year observation. Eur J Pediatr, 2003. 162(6): p. 421-5. [PubMed: 12756561]
- 25.
- Vuorio, A., et al., Statins for children with familial hypercholesterolemia. Cochrane Database Syst Rev, 2019. 2019(11). [PMC free article: PMC6836374] [PubMed: 31696945]
- 26.
- Manocha, D., et al., Safety profile of high-dose statin therapy in geriatric patients with stroke. South Med J, 2013. 106(12): p. 658-64. [PubMed: 24305522]
- 27.
- Ramsey, L.B., et al., PharmVar GeneFocus: SLCO1B1. Clin Pharmacol Ther, 2023. 113(4): p. 782-793. [PMC free article: PMC10900141] [PubMed: 35797228]
- 28.
- Kalliokoski, A. and M. Niemi, Impact of OATP transporters on pharmacokinetics. Br J Pharmacol, 2009. 158(3): p. 693-705. [PMC free article: PMC2765590] [PubMed: 19785645]
- 29.
- CPIC. SLCO1B1 allele functionality table 2021 12 Nov 2021 20 Nov 2023]; Available from: https://cpicpgx
.org/guidelines /cpic-guideline-for-statins /#:~:text=SLCO1B1 %20allele %20functionality%20table. - 30.
- Gaedigk, A., et al., Pharmacogene Variation Consortium: A Global Resource and Repository for Pharmacogene Variation. Clin Pharmacol Ther, 2021. 110(3): p. 542-545. [PMC free article: PMC8725060] [PubMed: 34091888]
- 31.
- PharmVar. SLCO1B1. 2023 26 Sep 2023 10 Dec 2023]; Available from: https://www
.pharmvar.org/gene/SLCO1B1. - 32.
- CPIC. SLCO1B1 diplotype-phenotype table. 2022 2022 20 Nov 2023]; Available from: https://files
.cpicpgx .org/data/report/current /diplotype_phenotype /SLCO1B1_Diplotype_Phenotype_Table .xlsx. - 33.
- Dashti, M., et al., Frequency of functional exonic single-nucleotide polymorphisms and haplotype distribution in the SLCO1B1 gene across genetic ancestry groups in the Qatari population. Sci Rep, 2022. 12(1): p. 14858. [PMC free article: PMC9437070] [PubMed: 36050458]
- 34.
- Saber-Ayad, M., et al., Statin-induced myopathy SLCO1B1 521T > C is associated with prediabetes, high body mass index and normal lipid profile in Emirati population. Diabetes Res Clin Pract, 2018. 139: p. 272-277. [PubMed: 29534995]
- 35.
- CPIC. SLCO1B1 frequency table 2022 11 Mar 2022 [cited 20 Nov 2023; Available from: https://cpicpgx
.org/guidelines /cpic-guideline-for-statins /#:~:text=SLCO1B1 %20frequency%20table. - 36.
- dbSNP [Cited 20 Feb 2024]. Available from https://www
.ncbi.nlm .nih.gov/snp/rs59502379. - 37.
- van de Steeg, E., et al., Complete OATP1B1 and OATP1B3 deficiency causes human Rotor syndrome by interrupting conjugated bilirubin reuptake into the liver. J Clin Invest, 2012. 122(2): p. 519-28. [PMC free article: PMC3266790] [PubMed: 22232210]
- 38.
- Memon, N., et al., Inherited disorders of bilirubin clearance. Pediatr Res, 2016. 79(3): p. 378-86. [PMC free article: PMC4821713] [PubMed: 26595536]
- 39.
- Simonson, S.G., et al., Rosuvastatin pharmacokinetics in heart transplant recipients administered an antirejection regimen including cyclosporine. Clin Pharmacol Ther, 2004. 76(2): p. 167-77. [PubMed: 15289793]
- 40.
- Park, J.W., et al., Pharmacokinetics and pharmacodynamics of fluvastatin in heart transplant recipients taking cyclosporine A. J Cardiovasc Pharmacol Ther, 2001. 6(4): p. 351-61. [PubMed: 11907637]
- 41.
- Newman, C.B., et al., Statin Safety and Associated Adverse Events: A Scientific Statement From the American Heart Association. Arterioscler Thromb Vasc Biol, 2019. 39(2): p. e38-e81. [PubMed: 30580575]
- 42.
- Wojtyniak, J.G., et al., Physiologically Based Precision Dosing Approach for Drug-Drug-Gene Interactions: A Simvastatin Network Analysis. Clin Pharmacol Ther, 2021. 109(1): p. 201-211. [PubMed: 33280091]
- 43.
- Wagner, J.B., et al., Impact of SLCO1B1 Genotype on Pediatric Simvastatin Acid Pharmacokinetics. J Clin Pharmacol, 2018. 58(6): p. 823-833. [PubMed: 29469964]
- 44.
- O'Brien, T.J., et al., Race and Drug Toxicity: A Study of Three Cardiovascular Drugs with Strong Pharmacogenetic Recommendations. J Pers Med, 2021. 11(11). [PMC free article: PMC8622254] [PubMed: 34834577]
- 45.
- Khine, H., et al., Statin-associated muscle symptoms and SLCO1B1 rs4149056 genotype in patients with familial hypercholesterolemia. Am Heart J, 2016. 179: p. 1-9. [PMC free article: PMC5014387] [PubMed: 27595674]
- 46.
- Cederberg, H., et al., Increased risk of diabetes with statin treatment is associated with impaired insulin sensitivity and insulin secretion: a 6 year follow-up study of the METSIM cohort. Diabetologia, 2015. 58(5): p. 1109-17. [PubMed: 25754552]
- 47.
- Laakso, M. and J. Kuusisto, Diabetes Secondary to Treatment with Statins. Curr Diab Rep, 2017. 17(2): p. 10. [PubMed: 28155189]
- 48.
- Laakso, M. and L. Fernandes Silva, Statins and risk of type 2 diabetes: mechanism and clinical implications. Front Endocrinol (Lausanne), 2023. 14: p. 1239335. [PMC free article: PMC10546337] [PubMed: 37795366]
- 49.
- Turkmen, D., et al., Statin treatment effectiveness and the SLCO1B1*5 reduced function genotype: Long-term outcomes in women and men. Br J Clin Pharmacol, 2022. 88(7): p. 3230-3240. [PMC free article: PMC9305522] [PubMed: 35083771]
- 50.
- Kitzmiller, J.P., et al., Candidate-Gene Study of Functional Polymorphisms in SLCO1B1 and CYP3A4/5 and the Cholesterol-Lowering Response to Simvastatin. Clin Transl Sci, 2017. 10(3): p. 172-177. [PMC free article: PMC5421731] [PubMed: 28482130]
- 51.
- Sivkov, A., et al., Relationship between genetic polymorphism of drug transporters and the efficacy of Rosuvastatin, atorvastatin and simvastatin in patients with hyperlipidemia. Lipids Health Dis, 2021. 20(1): p. 157. [PMC free article: PMC8573942] [PubMed: 34749751]
- 52.
- Wu, X., et al., Associations of the SLCO1B1 Polymorphisms With Hepatic Function, Baseline Lipid Levels, and Lipid-lowering Response to Simvastatin in Patients With Hyperlipidemia. Clin Appl Thromb Hemost, 2018. 24(9_suppl): p. 240S-247S. [PMC free article: PMC6714829] [PubMed: 30336686]
- 53.
- Generaux, G.T., et al., Impact of SLCO1B1 (OATP1B1) and ABCG2 (BCRP) genetic polymorphisms and inhibition on LDL-C lowering and myopathy of statins. Xenobiotica, 2011. 41(8): p. 639-51. [PubMed: 21425956]
- 54.
- Meyer zu Schwabedissen, H.E., et al., Function-impairing polymorphisms of the hepatic uptake transporter SLCO1B1 modify the therapeutic efficacy of statins in a population-based cohort. Pharmacogenet Genomics, 2015. 25(1): p. 8-18. [PubMed: 25379722]
- 55.
- Dou, Y., et al., Meta-Analysis of the SLCO1B1 c.521T>C Variant Reveals Slight Influence on the Lipid-Lowering Efficacy of Statins. Ann Lab Med, 2015. 35(3): p. 329-35. [PMC free article: PMC4390701] [PubMed: 25932441]
- 56.
- Kaewboonlert, N., et al., Lack of association between SLCO1B1 polymorphisms and lipid-lowering response to simvastatin therapy in Thai hypercholesterolaemic patients. J Clin Pharm Ther, 2018. 43(5): p. 647-655. [PubMed: 29575099]
- 57.
- Dagli-Hernandez, C., et al., Pharmacogenomics of statins: lipid response and other outcomes in Brazilian cohorts. Pharmacol Rep, 2022. 74(1): p. 47-66. [PubMed: 34403130]
- 58.
- PharmVar. CYP3A4. 2024 29 Jan 2024 16 Feb 2024]; Available from: https://www
.pharmvar.org/gene/CYP3A4. - 59.
- Elalem, E.G., et al., Association of cytochromes P450 3A4*22 and 3A5*3 genotypes and polymorphism with response to simvastatin in hypercholesterolemia patients. PLoS One, 2022. 17(7): p. e0260824. [PMC free article: PMC9286239] [PubMed: 35839255]
- 60.
- PharmVar. CYP3A5. 2024 20 Feb 2024]; Available from: https://www
.pharmvar.org/gene/CYP3A5. - 61.
- Melhem, A.L., et al., Common Statin Intolerance Variants in ABCB1 and LILRB5 Show Synergistic Effects on Statin Response: An Observational Study Using Electronic Health Records. Front Genet, 2021. 12: p. 713181. [PMC free article: PMC8517257] [PubMed: 34659336]
- 62.
- Wendt, F.R., et al., Biobank Scale Pharmacogenomics Informs the Genetic Underpinnings of Simvastatin Use. Clin Pharmacol Ther, 2021. 110(3): p. 777-785. [PMC free article: PMC8376807] [PubMed: 33837531]
- 63.
- Group, S.C., et al., SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med, 2008. 359(8): p. 789-99. [PubMed: 18650507]
- 64.
- Stewart, A., SLCO1B1 Polymorphisms and Statin-Induced Myopathy. PLoS Curr, 2013. 5. [PMC free article: PMC3871416] [PubMed: 24459608]
- 65.
- Carr, D.F., et al., Genomewide Association Study of Statin-Induced Myopathy in Patients Recruited Using the UK Clinical Practice Research Datalink. Clin Pharmacol Ther, 2019. 106(6): p. 1353-1361. [PMC free article: PMC6896237] [PubMed: 31220337]
- 66.
- Massmann, A., et al., SLCO1B1 gene-based clinical decision support reduces statin-associated muscle symptoms risk with simvastatin. Pharmacogenomics, 2023. 24(7): p. 399-409. [PMC free article: PMC10242433] [PubMed: 37232094]
- 67.
- Vassy, J.L., et al., Effect of Pharmacogenetic Testing for Statin Myopathy Risk vs Usual Care on Blood Cholesterol: A Randomized Clinical Trial. JAMA Netw Open, 2020. 3(12): p. e2027092. [PMC free article: PMC7716196] [PubMed: 33270123]
- 68.
- United States Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labeling. 2023 2 Feb 2024 20 Feb 2024]; Available from: https://www
.fda.gov/drugs /science-and-research-drugs /table-pharmacogenomic-biomarkers-drug-labeling. - 69.
- Elagolix [Cited 11 Dec 2023]. Available from https://go
.drugbank.com/drugs/DB11979. - 70.
- Viloxazine [Cited 20 Feb 2024]. Available from https://go
.drugbank.com/drugs/DB09185. - 71.
- Kalman, L.V., et al., Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther, 2016. 99(2): p. 172-85. [PMC free article: PMC4724253] [PubMed: 26479518]
Footnotes
- 1
The FDA has distinct labels for specific drug formulations. We have substituted the generic names for any drug labels in this excerpt. The FDA may not have labeled all formulations containing the generic drug. Certain terms, genes and genetic variants may be corrected in accordance with nomenclature standards, where necessary. We have given the full name of abbreviations, shown in square brackets, where necessary.
- Introduction
- Drug: Simvastatin
- Gene: SLCO1B1
- Linking SLCO1B1 Genetic Variation with Treatment Response
- Genetic Testing
- The SLCO1B1 Gene Interactions with Medications Used for Additional Indications
- Therapeutic Recommendations based on Genotype
- Nomenclature for Selected SLCO1B1 Alleles
- Acknowledgments
- References
- Effect of Pharmacogenetic Testing for Statin Myopathy Risk vs Usual Care on Blood Cholesterol: A Randomized Clinical Trial.[JAMA Netw Open. 2020]Effect of Pharmacogenetic Testing for Statin Myopathy Risk vs Usual Care on Blood Cholesterol: A Randomized Clinical Trial.Vassy JL, Gaziano JM, Green RC, Ferguson RE, Advani S, Miller SJ, Chun S, Hage AK, Seo SJ, Majahalme N, et al. JAMA Netw Open. 2020 Dec 1; 3(12):e2027092. Epub 2020 Dec 1.
- The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms.[Clin Pharmacol Ther. 2022]The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms.Cooper-DeHoff RM, Niemi M, Ramsey LB, Luzum JA, Tarkiainen EK, Straka RJ, Gong L, Tuteja S, Wilke RA, Wadelius M, et al. Clin Pharmacol Ther. 2022 May; 111(5):1007-1021. Epub 2022 Mar 11.
- SLCO1B1 variants and statin-induced myopathy--a genomewide study.[N Engl J Med. 2008]SLCO1B1 variants and statin-induced myopathy--a genomewide study.SEARCH Collaborative Group, Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. N Engl J Med. 2008 Aug 21; 359(8):789-99. Epub 2008 Jul 23.
- Review Pharmacokinetic comparison of the potential over-the-counter statins simvastatin, lovastatin, fluvastatin and pravastatin.[Clin Pharmacokinet. 2008]Review Pharmacokinetic comparison of the potential over-the-counter statins simvastatin, lovastatin, fluvastatin and pravastatin.Neuvonen PJ, Backman JT, Niemi M. Clin Pharmacokinet. 2008; 47(7):463-74.
- Review Lipid Screening in Childhood and Adolescence for Detection of Familial Hypercholesterolemia: A Systematic Evidence Review for the U.S. Preventive Services Task Force[ 2016]Review Lipid Screening in Childhood and Adolescence for Detection of Familial Hypercholesterolemia: A Systematic Evidence Review for the U.S. Preventive Services Task ForceLozano P, Henrikson NB, Dunn J, Morrison CC, Nguyen M, Blasi PR, Anderson ML, Whitlock E. 2016 Aug
- Simvastatin Therapy and SLCO1B1 Genotype - Medical Genetics SummariesSimvastatin Therapy and SLCO1B1 Genotype - Medical Genetics Summaries
Your browsing activity is empty.
Activity recording is turned off.
See more...

 1.
1.