NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Dean L, McEntyre J, editors. Coffee Break: Tutorials for NCBI Tools [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1999-.
The mitochondrion has cut back its genome substantially since taking up residence in cells as a symbiont 1.5 billion years ago, but it retains its personal transcription, translation and protein-assembling systems, including its tRNA genes. Even so, the mitochondrion is not fully self-sufficient — to varying extents yeast, plants and protozoan cells can borrow nuclear-encoded tRNA molecules to ease the task of translating transcripts of their mitochondrial genes. New data indicate that nuclear-encoded tRNAs can even be used to salvage errors in mitochondrial transcripts.
Figure
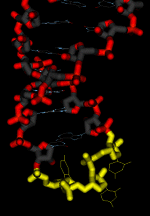
Tertiary structure of tRNA. The anticodon loop and stem of tRNALys is depicted in the image to the left. The three bases that compose the anticodon (in this example, "U-U-U") are highlighted in yellow.
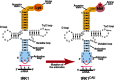
In the yeast Saccharomyces cerevisiae, only one tRNA (LysCUU) is carried into the mitochondrion, something it can do only if charged with an amino acid, and only if aided by cytosolic import factors. Among these factors is the precursor of the mitochondrial lysyl-tRNA synthetase (pre-MSK).
In a recent publication, researchers altered the aminoacylation identity of tRNALys CUU so that it was charged with methionine rather than lysine. Both in live yeast cells and in isolated mitochondria, the engineered tRNA could enter the mitochondrion, where the radiolabelled methionine charged on the imported tRNA was incorporated normally into mitochondrial proteins. A second, modified tRNALys version with alanine identity was also successfully used in vivo to suppress an amber (UAG) stop codon (a nonsense mutation) in the mitochondrial COX2 gene.
Defects in mitochondrial (mt) DNA, caused by base substitutions or rearrangements in genes that encode proteins or tRNAs underlie a range of human pathologies (as discussed in the previous highlight).
Could the technique used to modify mitochondrial mutations be adapted for use in humans, given that import of nuclear-encoded tRNAs into mammalian mitochondria has never been seen? It seems so, because isolated human mitochondria imported the yeast tRNALys CUU and its derivatives, provided that the human cytosolic extracts were supplemented with the yeast pre-MSK. The foreign tRNA was functional on the translational apparatus of human mitochondria, just as in yeast.
This recent innovation might be useful for replacing non-functional tRNAs or for suppressing nonsense mutations in mtDNA.
Story contributed by Tanita Casci, Nature Reviews Genetics
Search Organelle Genome Resources
Created: December 4, 2000
Click on the link below to start an html tutorial.
Evolution and the mitochondrial gene order of tRNAs
- Cytosolic help for mitochondrial defects - Coffee BreakCytosolic help for mitochondrial defects - Coffee Break
Your browsing activity is empty.
Activity recording is turned off.
See more...