NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
BioProject Help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2011-.
Introduction
The BioProject resource is a redesigned, expanded, replacement of the NCBI Genome Project resource. The redesign adds tracking of several data elements including more precise information about a project’s scope, material, and objectives. Genome Project identifiers are retained in the BioProject as the ID value for a record, and an Accession number has been added. Other changes include a more flexible approach to grouping projects and addition of data elements including fields for funding source and general relevance categories The web site presentation has been redesigned and some of the data elements that can be tracked in the new database design will be added to the public display once data has accumulated. In addition, database content is exchanged with other members of the International Nucleotide Sequence Database Collaboration (INSDC).
The BioProject database provides an organizational framework to access metadata about research projects and the data from those projects which is deposited, or planned for deposition, into archival databases maintained by members of the INSDC. The resource supports a variety of projects in terms of type and complexity, ranging from a focused genome sequencing project to a large international collaboration with multiple sub-projects such as sequencing, collecting genotype/phenotype data, calling sequence variants, or assaying epigenetic information. Data submitted to INSDC-associated databases cross-reference the BioProject identifier to support navigation between the Project and the project’s datasets. Therefore, the BioProject resource provides a reliable mechanism, for a variety of complex cases, to access specific datasets that can be difficult to find due to volume, sequential submissions over the course of a project, or submissions of distinct data types to multiple archival databases.
The definition of a set of related data, a ‘project’ is very flexible and supports the need to define a complex project and various distinct sub-projects using different parameters. For example, BioProject records can be established for:
- Genome sequencing and assembly
- Metagenomes
- Transcriptome sequencing and expression
- Targeted locus sequencing
- Genetic or RH Maps
- Epigenetics
- Phenotype or Genotype
- Variation detection
The database is not limited by taxonomy and as such includes information for studies of eukaryotes, prokaryotes, and environmental samples. Registration for a BioProject accession is encouraged for projects that result in a very large volume of data submissions, submissions from multiple members of a collaboration, or submissions to multiple archival databases. Registration for a BioProject accession is discouraged for small datasets for which the results are found in one (or a small number) of accession numbers such as a single viral or organelle genome sequencing study. A BioProject ID is required for some database submissions including dbVar, SRA, and GenBank microbial and eukaryotic genomes.
The database defines two types of projects: 1) Primary submission projects are directly associated with submitted data and may be registered by submitters of that data using the NCBI submission portal; 2) Umbrella projects reflect a higher-level organizational structure for larger initiatives or provide an additional level of data tracking that has been requested by a NIH institute. These projects are created by request, typically by a funding source. An Umbrella project may group projects that are part of a single collaborative effort but represent distinct studies that differ in methodology, sample material, or resulting data type. Complex studies may be represented with more than one layer of Umbrella project such that a highest-level Umbrella project is linked to one or more sub-project Umbrella projects which in turn are linked to one or more Primary submission projects that describe the data in more detail. As described below, the resource supports navigating up and down this hierarchy from any starting point within the hierarchy, as well as navigating to peer projects, e.g., those with a common Umbrella.
BioProject Quick Start Guide
BioProject records can be accessed by query, by browsing, or by following a link from another NCBI database.
Links to BioProject records may be found in several databases including dbVar, Gene, Genomes, GEO, and Nucleotide which includes GenBank or RefSeq nucleotide sequences for which there is a registered BioProject identifier.
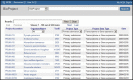
To access BioProjects by browsing, follow the link on the home page (Figure 1) to browse “By project attributes”. This page (Figure 2) supports browsing the database content by major organism groups, data attributes, or project data type. The table includes links to the NCBI Taxonomy database where additional information about the organism may be available and to the BioProject record where more information about the research project is reported.
The database can also be accessed by direct query. Queries can be entered from the BioProject home page, or by selecting the BioProject database name from the search menu found on the NCBI home page or other NCBI databases. The BioProject database can be queried using any text term or by restricting the query to a specific category using the Limits page or the Advanced Search page. Use the “Search Builder” menu available on the Advanced Search page to explore what fields are indexed and to build a complex Boolean query. Please refer to the Entrez Help chapter for a general description of Limits and Advanced Search pages.
Search Tips
You may search BioProject like any other NCBI database, namely by:
- Searching for an organism name
- Searching by the database accession or ID
- Searching for any word
- Restricting a search to a specific field using Limits
- Using the Advanced Search page to build a query restricted by multiple fields
The Limits page can be used to search by Project Type, Project Attributes, or Organism or Metagenome Groups.
See the Advanced Search page to explore the indexed fields and properties, view your search history, save search results, and view search details. Field restricted searching can be performed using the Search Builder. Restricted searches may also be entered manually by following the term with the name of the field in square brackets “[ ]”, as shown in the following examples. To see the indexed terms, choose a field from the pull down menu, then click on “Show index”.
Here are some representative searches:
| Find BioProjects by… | Search text example(s) |
|---|---|
| A species name | Saccharomyces cerevisiae[organism] |
| Project data type | "metagenome"[Project Data Type] |
| Project data type and Taxonomic Class | "transcriptome"[Project Data Type] AND Insecta[organism] |
| Publication | "10473380"[PMID] |
| Submitter organization, consortium, or center | JGI[Submitter Organization] |
| Sample scope and material used | "scope environment"[Properties] AND "material transcriptome"[Properties] |
| A BioProject database identifier | PRJNA33823 or PRJNA33823[bioproject] or 33823[uid] or 33823[bioproject] |
Display options
NCBI's Entrez system supports alternate display options for each of its databases. The options available can be browsed by clicking on Display Settings. The options presented in the Display Settings window depend on whether you are viewing a set of results, or just one record. In the former case, the Display settings window also provides choices for controlling the number of query results to display on the page.
BioProject offers four display settings, three of which are listed in the Display Settings menu, and the fourth is available by clicking on a record title. These display options are:
Summary
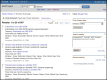
When you submit a query, the results are shown in the Summary format (or ‘docsum’) as shown in Figure 3. In the Summary format, each result is numbered, and a check box is provided at the left of the record. The check box enables you to select which of the records in the retrieval set that you want to review in another format, according to your selection in the Display Settings box. If none are checked, then all results are displayed in the selected format; this is the same as having all the boxes checked.
The text of the Summary includes the Project name or label (which is often the organism name), title, Taxonomy, Project data type, Attributes, the project source (if multiple, then only one is listed here), and the BioProject accession and ID. The Project name or label is linked to the full report page and the Taxonomy term is linked to NCBI’s taxonomy database.
Accessions List or BioProject ID List
These display only the BioProject accession number or the BioProject ID respectively, for the query result set.
Full Reports
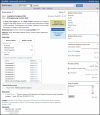
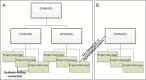
The full report page can be accessed by following the hyperlinked project name presented in the top row of each result returned in the Summary display. The Full Report display for Primary submission projects, as shown in Figure 4, includes the project name and/or title, a text description of the project (when provided), the project data type and specific project attributes, a project data section with data links, citations relevant to the project, taxonomic lineage, and information about the submitting group. Navigation tools are provided near the top of the report to facilitate navigation to NCBI’s taxonomically organized Genomes resource, ‘up’ to higher-level Umbrella projects, or ‘across’ to other BioProject records that are related by organism, or via a common Umbrella project. Umbrella project reports pages include a tabular report (when relevant) listing Umbrella sub-projects (see the HMP report PRJNA43021), or listing Primary submission projects, organized by Project data type, that it is grouping (Figure 5) (see the HMP Reference Genome project PRJNA28331). For all report pages, the right column provides navigation links and standard Entrez functions such as browsing history.
Some large initiatives are represented by more than one layer of umbrella projects (see Figure 6); for instance, a top-most level may identify the largest definition of the collaboration; a second level of umbrella projects identify the primary categories of data production; and finally a third layer represents the projects that actually generate the data that is submitted. The Human Microbiome project is an example of this type of complex hierarchy where the top-most project, PRJNA43021, represents the most inclusive definition of the initiative, and a secondary level (such as PRJNA28331) identifies a major sub-project to sequence multiple reference genomes each of which has a distinct project accession.
Genome sequencing projects may include a table that reports the accession numbers for assembled chromosomes, linkage groups, or other replicating molecules (such as organelles or plasmids), as well as the master accession for whole genome sequencing (WGS) projects.
When the experimental data for a BioProject is submitted to archival databases, it contains the BioProject accession which links the data to the BioProject. The Project Data table presents data counts from databases at NCBI that have links to the displayed BioProject record; if the displayed record is an Umbrella project, then the Data table presents a sum of data links for the grouped sub-projects. The counts are hyperlinked to the NCBI database indicated when a component (non-Umbrella) project is displayed. Hyperlinks are currently provided on Umbrella projects only in those cases when the database indicated holds links to a single component BioProject grouped under the Umbrella project; consequently, hyperlinks are not always present on Umbrella projects.
Submitting to BioProject
Projects can be registered with the BioProject database using the Submission portal (access the link from the home page). The Submission portal requires authentication and provides several login options, including National Institutes of Health eRA Login and other NIH logins. A login for the NCBI PDA (Primary Data Archives) system can be created here, if the user does not have any of the other types of accounts. Once logged in, the submission wizard provides a list of previously created submissions with some simple status information and a button to initiate a new submission. When making a new submission, the wizard presents a series of pages where information about the project can be entered. Required fields are marked with an asterisk (*) and simple validation identifies missing required content with red highlighting and warns of data attribute combinations that are observed less frequently. In-line help can be presented by hovering over the blue ‘?’ icons. The submission wizard pages must be completed in the order presented, but after that it is possible to navigate back to previous pages using tabs available along the top of the page. The content of each page is saved by clicking on the ‘Continue’ button located at the bottom of each page. To edit content on a previous page, the ‘Continue’ button must be clicked to save your changes. A submission may be started, set aside, and completed at a later time by signing back into the Submission wizard and selecting the incomplete project.
The page tabs presented by the Submission wizard are:
- Submitter – the name and email information is auto-filled if logging on using a NIH-based login approach and should identify the person who is entering the data in the form.
- General info – this page collects general descriptive information about the project, its relevance, whether it is part of a large initiative that has already registered with the BioProject resource, related web resources that are specific to the project, funding information, and information about the consortium or center name and/or data provider.
- Project data type – this page collects more specific information about the Sample Scope, Material, and other attributes (Capture and Method), as well as the Objective or goals of the project being registered. See the Glossary for descriptions of the attributes.
- Target – this page collects organism information (for projects focusing on an identified organism) or labeling information for projects that encompass multiple species whether identified or not (an environmental sample). An optional section, Biological Properties, collects general properties for samples that represent a single organism. This information is readily known for model organisms, but is not as readily available for lesser known organisms. Information about the number of chromosomes, genome size, mode of reproduction, and general habitat provides some useful context for other scientists who may be interested in the project and submitted data.
- Publications – this page collects publication information specific to the registered project. A publication identifier is required. A PubMed ID is preferred, but lacking that then a DOI may be supplied.
- Overview – this page presents a summary of the provided information. Click the ‘Submit’ button at the bottom of the page to complete the submission.
- BioProject Help - BioProject HelpBioProject Help - BioProject Help
Your browsing activity is empty.
Activity recording is turned off.
See more...

 1
1