From: The Database of Short Genetic Variation (dbSNP)

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
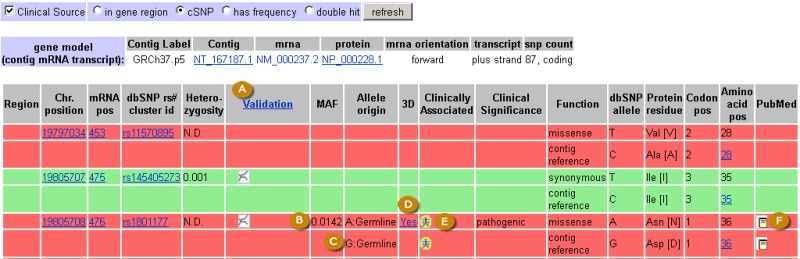
The refSNP Summary Report: The GeneView Display. The default GeneView display provides a tabular summary of variations mapped to splice variants of a particular gene. The variation summary is arranged in the sequential order that the variations appear in the genome and are color coded by functional class: in the example above, red indicates non-synonymous change and green indicates synonymous change. Additional colors not seen in the above example include white (in gene region), orange (UTR), blue (frame shift), yellow (intron). Attributes provided in the GeneView display include validation class (A), where definitions for graphic icons indicating validation class are viewed by clicking the “Validation” column header link (A); Minor Allele Frequency (if available) (B); allele origin (C) indicated as Germline or Somatic; a link to the Structure record (D) if 3D protein structure information is available; a link to the OMIM record (E) for those variations that have a clinical assertion; and a link to a list of Pubmed articles (F) that cite the variation.
From: The Database of Short Genetic Variation (dbSNP)

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.