From: The Database of Short Genetic Variation (dbSNP)

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
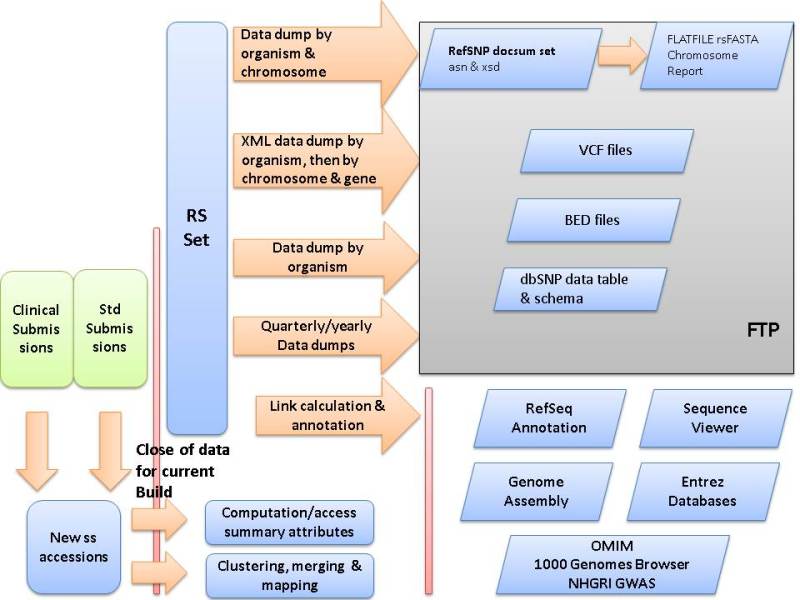
SNP Build Cycle. The dbSNP build cycle starts with close of data for new submissions. dbSNP calculates summary attributes and provide submitter asserted summary attributes for each refSNP cluster. These attributes include genotype, false positive status, minor allele frequency (MAF), asserted allele origin, asserted clinical significance, and many others. dbSNP maps all data, including existing refSNP clusters and new submissions, to the available reference genome sequence for the organism. If no genome sequence is available, dbSNP maps the data to non-redundant DNA sequences from GenBank. dbSNP uses map data on co-occurrence of hit locations to either merge submissions into existing clusters or to create new clusters. dbSNP then annotates the new non-redundant refSNP (rs) set on reference sequences and dump the contents of dbSNP into a variety of comprehensive formats on the dbSNP FTP site for release with the online build of the database. dbSNP then creates links from each record to internal and external resources that can provide additional data for each record.
From: The Database of Short Genetic Variation (dbSNP)

NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.