This publication is provided for historical reference only and the information may be out of date.
Scope
Bookshelf is a biomedical literature trove, whether you are preparing for a college biology test, studying health trends, or investigating the molecular basis of a gene mutation. Bookshelf (http://www.ncbi.nlm.nih.gov/books/ ) is an online resource providing free access to the full text of books and documents in life sciences and health care, built and maintained by the National Center for Biotechnology Information (NCBI) within the National Library of Medicine (NLM) (1). Bookshelf includes books, reports, documentation, and databases in life sciences and health care.
Bookshelf data is tagged in XML in the NCBI Book DTD (Document Type Definition), which is modeled after the NLM Journal Article DTDs. Book content follows a processing route similar to journal articles; tagging book data in a format similar to journal articles in PMC (PubMed Central) has enabled Bookshelf to use existing PMC infrastructure and workflows for processing book content.
Bookshelf aims to further advance science and improve health care through the collection, exchange, and dissemination of books and related documents in life sciences and health care. As a literature resource at NCBI, Bookshelf serves to provide annotations for the factual information residing in the genomic and molecular databases such as Gene and PubChem, and facilitate the discovery of this information.
History
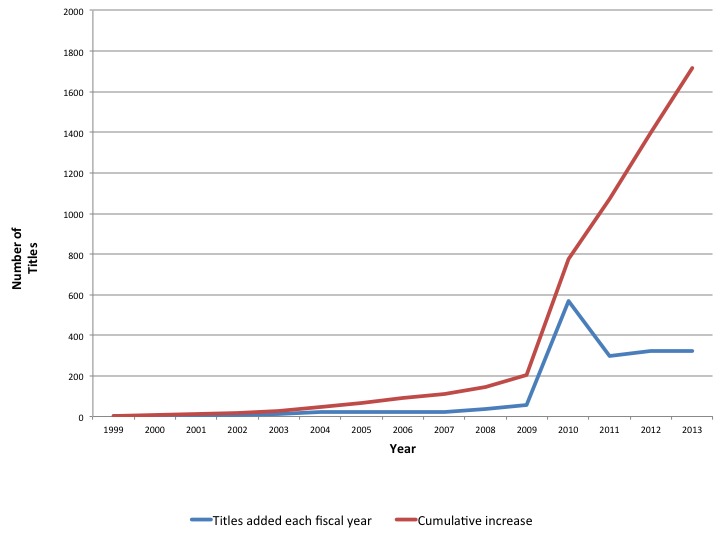
Bookshelf started in 1999, with a single book, the third edition of Molecular Biology of the Cell, Alberts et al. (2). The first few books in Bookshelf were college text books. In the early days of Bookshelf, terms in PubMed abstracts were linked to the books which served as encyclopedic references for these terms. With the introduction of the Health Services/Technology Assessment Texts (HSTAT) collection to Bookshelf in 2004, a large number of health reports were added to Bookshelf. Today, there are over 1700 titles in Bookshelf (see Figure 1).
Figure 1.
Number of titles added in Bookshelf each fiscal year (October to September) and cumulative growth. The spike in 2010 represents a restructuring of the HSTAT collection.
The Collection and Content
The collection is broadly biomedical in scope, comprising a diversity of works. They include books, reports, literature databases, and documentation ranging from basic undergraduate text books to specialized publications in life sciences and healthcare. Titles are selected for the collection based on three criteria: (1) scope, as defined by NLM's Collection Development Guidelines; (2) scientific and editorial quality of the content; and (3) technical considerations, such as the quality of submitted files. Some works are in the public domain, whereas others are copyrighted works for which the copyright holders have granted NCBI distribution rights. Once content is selected for the collection, participants sign an agreement. See Information for Authors and Publishers for details on the selection process, how to apply, and to view the agreement.
Bookshelf serves users seeking biomedical information; they include college and graduate students, scientists, healthcare professionals, and patients. The free availability of the content ensures that the information is accessible by users who might otherwise not have access to this data. The content providers agree to make the content freely available; they include authors, editors, publishers, and administrators from universities, publishing houses, US and international government agencies, as well as organizations in the health sector. Publishers and content providers also benefit when their content is widely distributed to the general public, to health care professionals, and to a population of students who will become the next generation of biomedical researchers, clinicians, and teachers.
Some content providers also agree to participate in the Open Access subset. For content in the Open Access subset, XML, image and supplementary files are shared, allowing for redistribution and reuse of the content.
Data Model
Format and Structure
Early in the project, Bookshelf used a DTD based on the ISO 12083 article DTD for tagging data in XML format. As the project grew with more data being added, the tag set had to be modified, complicating data management and rendering. This led to the development of the NCBI Book DTD, which is modeled along the same design principles as the DTDs of the Journal Article Tag Suite (JATS), and utilizes many of same modules. Bookshelf XML data are currently tagged in the NCBI Book DTD, v2.3. The similarities between book chapters and journal articles, and between their shared tag sets, have permitted Bookshelf to leverage the robust PMC architectural framework as well as existing PMC workflows and tools for handling the data. The NCBI Book DTD in the context of JATS has been discussed in detail (3).
Submission, XML Conversion, and Storage
Tagging content semantically in XML is one of the most complex and costly operations for Bookshelf. To enable continued maintenance of the corpus of book data, and continued growth of Bookshelf, it has been necessary to balance the needs of the publisher with the resources of the Bookshelf by streamlining the number of submission formats. To this end, Bookshelf recently moved toward a requirement for data submission in semantically tagged XML, which permits partial or complete automation of data processing. XML data are submitted either in the NCBI Book DTD or in an alternate DTD (e.g., DocBook). When submission utilizes an alternate DTD, Bookshelf employs XSLT converters to transform the XML to the NCBI Book DTD format. For submission of data in NCBI Book DTD XML, tagging guidelines have been developed and are based on similar tagging guidelines for PMC. These guidelines are intended to guide proper tagging practice through tagged samples, to reduce the variability in tagging data elements, and to facilitate data exchange.
A subset of Bookshelf projects that require frequent updates are authored in a specialized Microsoft Word template that utilizes styles to semantically tag the document elements, such as the title, author list, etc. The documents are converted to XML using the in-house NCBI Word Converter tool that utilizes the eXtyles product (Inera, Inc.) for reference processing. Documents are updated in Microsoft Word and reprocessed using the Word Converter. Legacy projects involving print publications are submitted in PDF format and are converted by third-party vendors to NCBI Book DTD XML. FTP is the main portal for data submission.
For the majority of books (>99.5%), XML, image, source files (example, publisher-supplied PDFs, Word), and supplementary files are stored in a content management system (CMS), built in-house for the Bookshelf project. The CMS is the destination hub for NCBI Book DTD XML data that is received through a number of workflows and the staging area for ingest and subsequent processing of book data. All XML content stored in the CMS is in the form of a master XML document that describes the book’s metadata and individual book part elements such as chapters and appendices. For convenience in editing the book, the individual book chapters and appendices are in separate XML files. Support data files for the book such as figure images, PDFs, supplementary files, as well as original source files are also stored in CMS. In the CMS, book data are checked for validation against the DTD, conformance to an in-house stylechecker (which runs additional checks beyond XML validation to ensure data quality), and additional integrity checks to ensure that all files associated with the book are available (see below, Performing Quality Assurance).
The different operations such as validation, style check, integrity check and loading to PMC can be selected and run separately by the user. However, these operations can also be defined as a workflow and the workflow can be run as an interactive or batch process that ensures that the operations are executed in the intended order specified in the workflow. The workflow is described as an XML document. The elements of the workflow are described using W3C schema and include the CMS operations and conditional and branching logic to execute the next step dependent on the success of previous steps. Defining workflows using XML gives users the flexibility of creating custom workflows and modifying them as future needs change.
The CMS is set up so that most operations dealing with the content processing can be done or initiated from the CMS. For example, an XML file can be edited by selecting it from CMS and running the Oxygen XML Editor (SyncRO Soft SRL) and saving the results back to CMS. There is no need to copy the file outside CMS and edit it separately and then upload the edited file to CMS. Another example is authoring content using the Microsoft Word template (above). There is a separate area in the CMS for storing book chapters authored in MS Word. These Word documents can be converted to XML by initiating the conversion action from CMS. The Word documents are converted to XML and the results stored in CMS.
Books contents in CMS can be searched using XQuery. The XQuery scripts can be stored and edited directly in CMS and run against any set of books. The XQuery and workflows can be set to run immediately or at a future time using a built-in scheduler. This enables workflows and queries that require heavy processing to be performed at times when the system is not so heavily used.
Dataflow
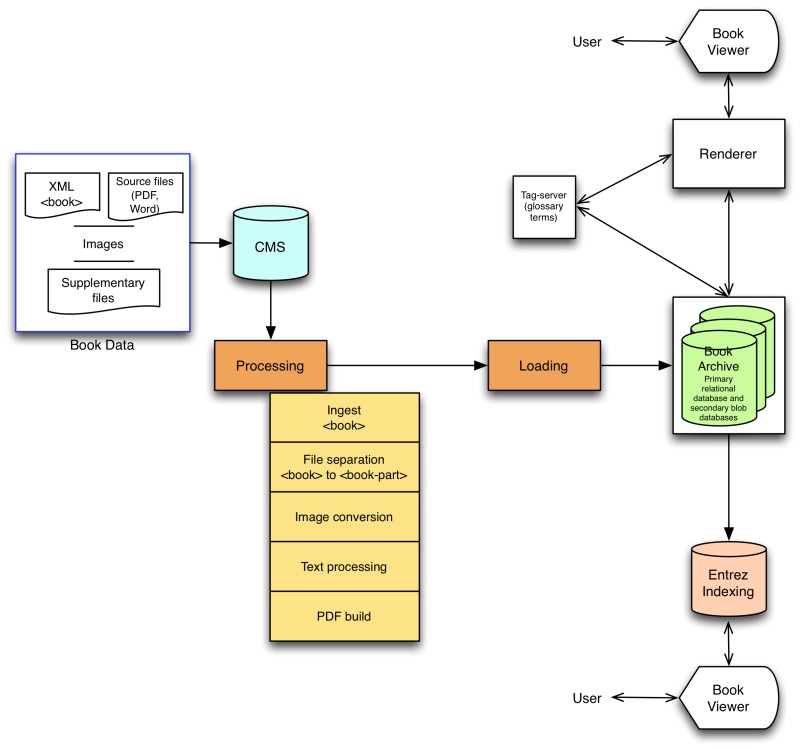
From the CMS, content is then processed for storage in the books archive to enable fast delivery to the Web, and for the automated creation of alternative formats (example, PDF). The main steps of data processing are: (a) ingest, (b) “chop-it-up” process, (c) text and image processing, and (d) PDF build (see Figure 2). Ingest begins with downloading XML, image, and supplementary files from the CMS onto the file system then bundling them to create a tar file; in cases where the CMS is bypassed (<0.5% of books), data is directly ingested following deposit to the FTP site.
Figure 2.
Books data workflow.
Chop-it-up and text processing involve XSLT transformation on XML data, creating XML output. During the chop-it-up process, the single independently validating NCBI Book DTD XML document with root element <book> is separated into independently validating XML documents with root element <book-part>; i.e., the book is divided into standalone book units such as front-matter sections, chapters, appendices, or reference lists. Book-metadata is carried into every book part. The creation of article-like <book-part> XML files from the <book> XML has provided the basis for using the PMC workflows and tools for Bookshelf data processing.
Text processing and image conversion occur in parallel. For text conversion, the software resolves named entities, handles special or custom characters and custom math, validates XML, and runs the stylechecker. For image conversion, the software which runs on open-source ImageMagick (ImageMagick Studio) determines image dimensions and properties, such as size, type, and resolution, resizes images per Bookshelf specifications, and creates for each image a thumbnail, a Web-resolution JPEG file, and a high-resolution JPEG file (if the source files were of high resolution).
PDFs are created for book chapters if not provided by the content provider and if their creation and display in Bookshelf is permitted. The PDF build software uses the XML output of text conversion and creates a formatting object (FO) file, gathers image heuristics, and resizes images so they are compatible with print layout. The Antenna House formatter (Antenna House, Inc.) creates the PDF from the formatting object file.
Loading to the Database
The loading software identifies the XML files for addition or replacement and loads them to the database. Each book in the database is referred to as a domain. The loader validates the data, and performs checks for file types and associated files; resolves loading of files associated with each XML file, such as images, equations, multimedia, and supplementary files. It parses the XML for key metadata information, such as book-part identifiers for storage in the main database tables. Citations that have PubMed identifiers are stored in the database. The loader creates a unique accession ID, with the “NBK” prefix for each book part.
The book database is very similar in design to the PMC article database (see SQL Databases). It is actually a database cluster with a primary database for the main relational tables holding book and book part information, as well as their properties and attributes; and several secondary blob databases for holding the XML and associated file blobs.
Rendering
Bookshelf dynamically renders the book-part XML to HTML Web pages at request time. The architecture closely corresponds to the PubMed Central (PMC) rendering model: The NCBI frontend system analyzes a request from a client browser and routes it to the renderer, a FastCGI program written in C++. The program retrieves the book-part XML as well as additional information about the book-part, for example PubMed IDs of references cited in the content. It runs the data through an XSLT transformation and then passes it back to the frontend, which returns an HTML page to the client. Bookshelf uses the PMC Caching system in order to deliver its pages faster. It also exploits the PMC TagServer as a tool to enrich the content, for example by mining and storing glossary terms mentioned across a book-part.
Performing Quality Assurance
Quality assurance checks aim to protect the fidelity of data through all stages of processing and ensure accurate rendering and retrieval by the user. Bookshelf uses both manual and automated procedures for performing quality assurance checks. Metadata checks against the source documents, as well as integrity checks (to ensure that all book files are included) are performed in the CMS. Following ingest, processing and loading to the SQL databases, checks are also performed in the Book Viewer application to ensure that all data is accurately rendered.
Indexing
Bookshelf records are indexed in Entrez, NCBI’s global indexing, retrieval, and discovery system. Entrez records are created for a complete book, for its individual chapters as well as for lower-level units, such as sections or tables. A Bookshelf Entrez record mainly contains:
- Main search text which comprises the body of the content unit;
- Search fields based on bibliographic and subject metadata, for example, authors or title; and
- Specially computed keywords and phrases.
The indexing process runs each night. A Perl program retrieves the book part XML files from the database. It passes it through an XSLT transform to produce simplified “indexing documents,” extracting the bibliographic search fields and the search text. It also interfaces with a program maintained by NCBI’s Computational Biology Branch to compute important keywords from the book XML and merges those into the indexing document. The latter is then fed into the global Entrez indexing pipeline.
In addition to the main indexing records, the process also produces Entrez filters and links: it collects, for example, all records belonging to a particular bibliographic series or set into a filter, which enables the user to limit her or his search to a particular collection of interest. It creates link pairings to other NCBI databases, for example, to PubMed Records cited in a chapter or to a Gene records tagged in the book XML.
Access
Search
Users can search Bookshelf for a term or phrase across all books or in a single book. An advanced search builder and the ability to apply limits to the search query are available. Standard search features familiar to PubMed users, such as Save search, Send to Clipboard, and Search details are also available. See Searching Bookshelf for details on performing a Bookshelf search.
Example
Search for term: heart attack
Bookshelf uses some of the query processing facilities available in the Entrez system. Search terms, for example, are expanded via a Medical Subject Headings (MeSH) translation table used also in PubMed. Similarly, the system employs a spell-checker or uses phrase tokenization if an original user query yields no results.
Browse
Books can be browsed using an application that allows users to filter the list of books by entering a term into a text box or by selection of one or more of the following categories: subject, type of publication, and publisher. A URL request is sent from the client to the browse application backend and the backend response is handled using AJAX (Asynchronous JavaScript and XML), allowing fast loading of the page without a reload. This tool is available at: http://www.ncbi.nlm.nih.gov/books/browse/ . See Browsing Bookshelf for details on using the browse tool.
Read
The book viewer application presents book content to the reader, as in the page you are currently reading. It facilitates navigation within the book, as well as within the page. Through this application, users can access all features of the book such as tables, figures, glossaries, bibliographic reference lists, download alternate formats, view bibliographic information, copyright and permissions, and cite the content.
Related Resources
A subset of books and book chapters in Bookshelf are indexed in PubMed. They are searchable using the filters “pmcbook” and “pmcbookchapter” respectively in PubMed. They are identifiable in the PubMed result summary by the label “Books and Documents.” MARC records are available for Bookshelf titles and can be downloaded from the following FTP site: ftp://ftp.ncbi.nlm.nih.gov/pub/bookshelf/. Bookshelf catalog records can also be found in the NLM Catalog.
References
- 1.
- Hoeppner MA. NCBI Bookshelf: books and documents in life sciences and health care. Nucleic Acids Res. 2013 Jan;41(Database issue):D1251–60. Epub 2012 Nov 29PubMed Central PMCID: PMC3531209; doi. [PMC free article: PMC3531209] [PubMed: 23203889] [CrossRef]
- 2.
- Alberts B, Bray D, Lewis J, et al. Molecular Biology of the Cell. 3rd edition. New York: Garland Science; 1994. Available from: http://www
.ncbi.nlm.nih .gov/books/NBK20684/ - 3.
- Latterner M, Hoeppner M. Bookshelf: Leafing through XML. 2010 Oct 12. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2010 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-. Available from: http://www
.ncbi.nlm.nih .gov/books/NBK47113/
Publication Details
Author Information and Affiliations
Authors
Marilu Hoeppner, 1 Martin Latterner,1 and Karanjit Siyan1.
1 Martin Latterner,1 and Karanjit Siyan1.Affiliations
 Corresponding author.
Corresponding author.Publication History
Created: March 18, 2013; Last Update: November 4, 2013.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Hoeppner M, Latterner M, Siyan K. Bookshelf. 2013 Mar 18 [Updated 2013 Nov 4]. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.