This publication is provided for historical reference only and the information may be out of date.
Scope
The BioSample database (1) stores submitter-supplied descriptive information, or metadata, about the biological materials from which data stored in NCBI’s primary data archives are derived. NCBI’s archives host data from diverse types of samples from any species, so the BioSample database is similarly diverse; typical examples of a BioSample include a cell line, a primary tissue biopsy, an individual organism or an environmental isolate.
The BioSample database promotes the use of structured and consistent attribute names and values that describe what the samples are as well as information about their provenance, where appropriate. This information is important for providing context to the derived data so that it may be more fully understood; it adds value, promotes re-use, and enables aggregation and integration of disparate data sets, ultimately facilitating novel insights and discoveries across a wide range of biological fields.
BioSample records are indexed and searchable. They are also reciprocally linked with the BioProjects in which they participate, as well as with derived experimental data in NCBI’s primary archives including Sequence Read Archive (SRA), Gene Expression Omnibus (GEO), database of Genotypes and Phentotypes (dbGaP), as well as sections of GenBank, including Expressed Sequence Tags (EST), Genome Survey Sequences (GSS), Whole Genome Shotgun (WGS), and Transcriptome Shotgun Assembly (TSA) sequences.
In summary, the BioSample database provides a dedicated environment in which to:
- Capture sample metadata in a structured way by promoting use of controlled vocabularies for sample attribute field names.
- Link sample metadata to corresponding experimental data across multiple archival databases.
- Reduce submitter burden by enabling one-time upload of a sample description, then referencing that sample as appropriate when making data deposits to other archives.
- Support cross-database queries by sample description.
History
As the number and complexity of primary data archives supported by NCBI expands, a need has emerged for a shared database in which to host information about the biological samples from which those data are derived. Historically, each archive developed its own conventions for collecting sample metadata, with limited standardization of descriptions and no mechanism to indicate when the same sample was used across multiple sets of data. Furthermore, there is a growing awareness in the research community that sample metadata is essential for interpreting the data itself, and that opportunities for data re-use, aggregation, and integration increase with improved metadata.
The BioSample database was launched in 2011 to begin to help address these needs. It facilitates the capture and management of structured metadata descriptions for diverse biological samples and encourages data producers to provide a rich set of contextual metadata with their data submissions. The database was initially populated with existing sample descriptions extracted from SRA, dbGaP, EST and GSS. Over time, more NCBI archives are moving towards requiring BioSample deposit as part of data submission. As of May 2013, the database hosts almost 2 million BioSample records encompassing 18,000 species.
Data Model
The BioSample database stores descriptions of the biological materials used to generate data hosted by any of NCBI’s primary data archives and, consequently, are very heterogeneous in nature. This, together with the fact that the content and granularity of metadata submitted to NCBI tends to be dependent on the context of the study, presents significant challenges in terms of procuring consistent sample descriptions from submitters.
To help address these challenges, the BioSample Submission Portal guides submitters into providing appropriate information. A number of common BioSample types are defined in the database, each comprising a package of relevant attributes with which to describe the sample. By guiding and encouraging submitters to use such attribute packages, it can be expected that the descriptions for samples deposited via this route will converge and become more consistent over time.
The full list and definitions of BioSample types and attributes is available for preview and download. Examples include “Pathogen affecting public health,” which is intended to procure information considered useful for the rapid analysis and trace back of pathogen samples, and the MiXS minimum information checklists as developed by the Genomics Standards Consortium (2) that are intended for standardizing descriptions of samples from which genomes, metagenomes, and targeted locus sequences are derived.
Attributes define the material under investigation using structured name: value pairs, for example:
tissue: liver
collection date: 31-Jan-2013
After specifying the sample type, the user is presented with a list of required and optional attribute fields to fill in, as well as the opportunity to supply any number of custom descriptive attributes. For example, if a submitter specifies that their sample is a clinical pathogen, they are required to input information about collection locality and date, host and isolation source. In addition, submitters are encouraged to provide information for additional attributes that further describe the host, disease state, etc. The values provided in some fields undergo validation to ensure proper content or format. The BioSample database is extendible in that new types and attributes can be added as new standards develop.
In addition to BioSample type (called Model in the schema) and attributes, each BioSample record also contains:
IDs: An identifier block that lists not only the BioSample accession assigned to that record, but also any other external sample identifier, such as that issued by the source database or repository.
Organism: The organism name and taxonomy identifier. The full taxonomic tree is displayed and searchable.
Title: BioSample title. A title is auto-generated if one is not supplied by the submitter.
Description: [optional] A free text field in which to store non-structured information about the sample.
Links: [optional] URL to link to relevant information on external sites.
Owner: Submitter information, including name and affiliation where available.
Dates: Information about when the record was submitted, released, and last updated.
Access: Statement about whether the record is fully public or controlled access (that is, in dbGaP).
BioSample records of interest include:
Reference BioSamples: While many samples can be considered unique and are used only once, other samples, including commercial cell lines or bacterial isolates, are used repeatedly by the research community. Major vendors, including the American Type Culture Collection (ATCC), the Coriell Institute for Medical Research , and the Leibniz Institute German Collection of Microrganisms and Cell Cultures (DSMZ), are working with us to generate official representations of commonly used and highly referenced samples. These are flagged as Reference BioSamples, so submitters who use these samples may bypass BioSample submission and simply reference relevant Reference BioSample records when depositing experimental data in any of NCBI's primary data archives. Also, efforts are underway to map existing data from across NCBI archives to Reference BioSample records. Consequently, these Reference BioSample records serve as hubs from which users can quickly locate a multitude of diverse data sets and projects derived from a given sample.
Clinical samples: The BioSample database does not support controlled access mechanisms and thus cannot host human clinical samples that may have associated privacy concerns. Instead, clinical samples continue to be deposited in NCBI's dbGaP database. The dbGaP database then deposits abridged BioSample records that have had sensitive data attributes removed. This allows users to locate these data in BioSample, and then apply to dbGaP for access to the full descriptions as necessary.
Authenticated human cell line samples: The BioSample database hosts a growing collection of authenticated human cell line records aimed at addressing the problem of cell line misidentification (3). These records contain verified STR (short-tandem-repeat) profile information and supporting electopherogram evidence which researchers can use as a reference when checking the authenticity and purity of the cell lines from which they are publishing data.
Dataflow
Researchers typically initiate a deposit to BioSample as part of a submission to one of NCBI’s primary data archives, and usually before a manuscript describing the data has been submitted to a journal for review. Researchers use their NCBI account to login and register BioSample submissions using a Web-based Submission Portal that guides them through a series of forms for input of metadata describing their samples. An XML–based submission route is also available for frequent submitters. In addition, direct data deposits to dbGaP and GEO trigger automatic creation of BioSample records.
The BioSample Submission Portal enforces provision of a minimal set of metadata via mandatory attributes for specific sample types, as well as encourages rich metadata by supporting the provision of any number of custom attributes. But ultimately, BioSample is a submitter-driven repository in that submitters are responsible for the quality and content of their deposits. Database staff respond to queries and report errors but, as with other primary data archives, submitted data are not subject to extensive curation. After passing syntactic validation, each sample is assigned a BioSample accession number which has prefix SAMN, e.g., SAMN02048828. This accession number can subsequently be referenced as appropriate when submitting corresponding experimental data to the archival databases.
The BioSample records are typically released in conjunction with corresponding experimental data. At that time, the BioSample records are loaded and indexed in the BioSample database that is part of NCBI's Entrez search and retrieval system, where they may be queried and downloaded. The records are reciprocally linked to other databases where appropriate, including SRA, dbGaP, GEO, GenBank and BioProject, facilitating easy navigation to derived and related data.
Access
BioSample records may be accessed by query or by following a link from another NCBI database.
Query: Effective searches may be accomplished using the search box on the BioSample home page. As with other NCBI Entrez databases, a simple free text keyword search is often sufficient to locate relevant data. However, BioSample data are indexed under several fields, meaning that users can refine their search by constructing fielded queries. Some example fielded queries are listed below and include searching by organism, attribute, or package. Users can write and execute their own search statements directly in the search boxes or use the Advanced search page to explore the indexed fields and construct multi-part fielded search statements. The Limits page may be used to restrict retrievals according to access-level, source databases, and publication dates.
Download: BioSample record content may be downloaded using the Send to: feature on the search results pages that allows download of individual or batch BioSample retrievals in text or XML formats. Furthermore, programmatic query and download functions are available using Entrez Utilities.
Linking: BioSample records are reciprocally linked to related records in the archival databases. This allows users to link to, e.g., corresponding genome assembly records in the Nucleotide database, or raw sequence reads in SRA, or to navigate to the BioProject(s) in which the sample participates.
Example queries
Retrieve pathogen BioSamples released in the first quarter of 2013
package pathogen[Properties] AND 2013/1:2013/3[Publication date]
Retrieve BioSamples derived from bacteria of genus Shigella and for which SRA data is available:
shigella[organism] AND biosample sra[filter]
Retrieve BioSamples that conform to the MIGS/MIMS/MIMARKS.water package:
package migs/mims/mimarks water[Properties]
Retrieve BioSamples derived from mouse and for which strain and age information is available:
(strain[Attribute Name] AND age[Attribute Name]) Mus musculus[organism]
Retrieve BioSamples derived from fibroblast cells:
cell type fibroblast[Attribute]
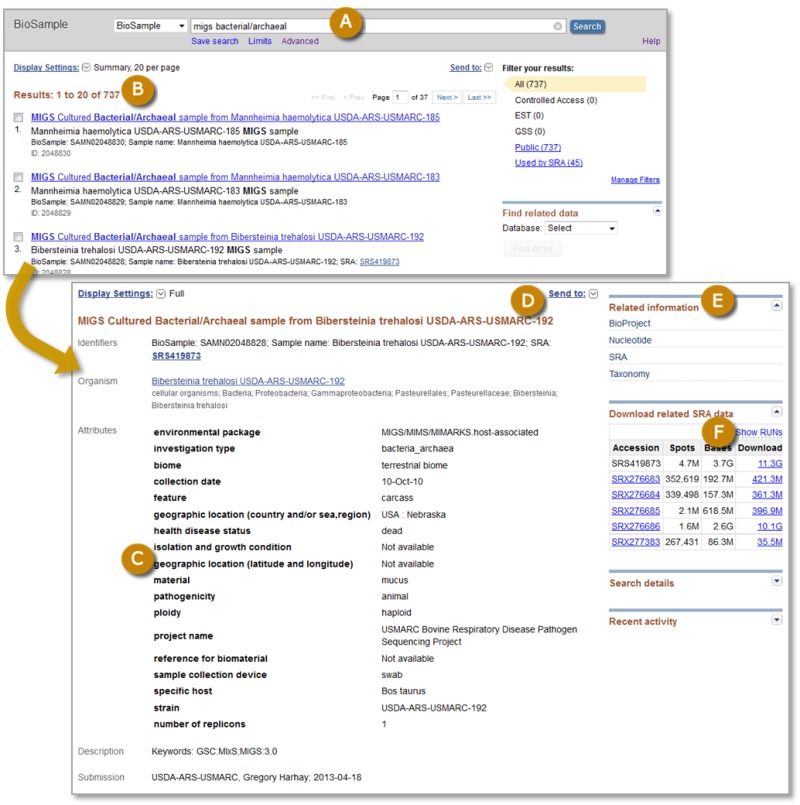
Figure 1.
Screenshots of BioSample search results (top panel) and a full BioSample record (bottom panel). Users enter a query into the Search box, or use the Limits or Advanced search pages (A) and retrieve a list of matching BioSamples (B). Search results are displayed in Summary format by default, which presents the title, organism, sample type, and identifiers. Clicking a title takes the user to the full record that lists all the sample attributes, identifiers, and submitter information (C). The Send to: feature (D) allows download of individual or batch BioSample retrievals in text or XML formats. Links are provided to related records in other archives (E), in this case BioProject, Nucleotide, SRA, and Taxonomy. Where appropriate, an option to download SRA sequence data generated from that sample is provided (F).
References
- 1.
- Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, Yaschenko E, Ostell J. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res 2012Jan 40Database issueD57–63. [PMC free article: PMC3245069] [PubMed: 22139929]
- 2.
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, Vaughan R, Hunter C, Park J, Morrison N, Rocca-Serra P, Sterk P, Arumugam M, Bailey M, Baumgartner L, Birren BW, Blaser MJ, Bonazzi V, Booth T, Bork P, Bushman FD, Buttigieg PL, Chain PS, Charlson E, Costello EK, Huot-Creasy H, Dawyndt P, DeSantis T, Fierer N, Fuhrman JA, Gallery RE, Gevers D, Gibbs RA, San Gil I, Gonzalez A, Gordon JI, Guralnick R, Hankeln W, Highlander S, Hugenholtz P, Jansson J, Kau AL, Kelley ST, Kennedy J, Knights D, Koren O, Kuczynski J, Kyrpides N, Larsen R, Lauber CL, Legg T, Ley RE, Lozupone CA, Ludwig W, Lyons D, Maguire E, Methe BA, Meyer F, Muegge B, Nakielny S, Nelson KE, Nemergut D, Neufeld JD, Newbold LK, Oliver AE, Pace NR, Palanisamy G, Peplies J, Petrosino J, Proctor L, Pruesse E, Quast C, Raes J, Ratnasingham S, Ravel J, Relman DA, Assunta-Sansone S, Schloss PD, Schriml L, Sinha R, Smith MI, Sodergren E, Spo A, Stombaugh J, Tiedje JM, Ward DV, Weinstock GM, Wendel D, White O, Whiteley A, Wilke A, Wortman JR, Yatsunenko T, Glockner FO. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol 2011May 29(5):415–20. [PMC free article: PMC3367316] [PubMed: 21552244]
- 3.
- Masters JR. Cell-line authentication: End the scandal of false cell lines. Nature 2012Dec 13 492(7428):186. [PubMed: 23235867]
Publication Details
Author Information and Affiliations
Authors
Tanya Barrett, PhD1.Affiliations
Publication History
Created: November 14, 2013.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Barrett T. BioSample. 2013 Nov 14. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-.