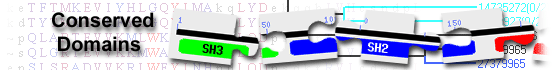Conserved domains on
[gi|50812102|ref|YP_054501|]
View


COX1 intron 3 ORF (mitochondrion) [Kluyveromyces lactis]
LAGLIDADG family homing endonuclease( domain architecture ID 10469810)
LAGLIDADG family homing endonuclease belongs to a large family of homing endonucleases that each contain one or two copies of a motif that resembles the consensus sequence LAGLIDADG and is directly involved in the DNA cutting process
List of domain hits

| Name | Accession | Description | Interval | E-value | |||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
308-394 | 1.91e-04 | |||
LAGLIDADG endonuclease; : Pssm-ID: 395767 Cd Length: 101 Bit Score: 40.31 E-value: 1.91e-04
|
|||||||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
142-233 | 6.04e-04 | |||
LAGLIDADG endonuclease; : Pssm-ID: 395767 Cd Length: 101 Bit Score: 38.77 E-value: 6.04e-04
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
308-394 | 1.91e-04 | |||
LAGLIDADG endonuclease; Pssm-ID: 395767 Cd Length: 101 Bit Score: 40.31 E-value: 1.91e-04
|
|||||||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
142-233 | 6.04e-04 | |||
LAGLIDADG endonuclease; Pssm-ID: 395767 Cd Length: 101 Bit Score: 38.77 E-value: 6.04e-04
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
308-394 | 1.91e-04 | |||
LAGLIDADG endonuclease; Pssm-ID: 395767 Cd Length: 101 Bit Score: 40.31 E-value: 1.91e-04
|
|||||||
| LAGLIDADG_1 | pfam00961 | LAGLIDADG endonuclease; |
142-233 | 6.04e-04 | |||
LAGLIDADG endonuclease; Pssm-ID: 395767 Cd Length: 101 Bit Score: 38.77 E-value: 6.04e-04
|
|||||||
| Blast search parameters | ||||
|