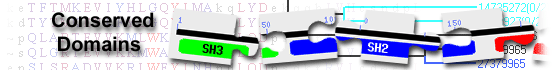|
Name |
Accession |
Description |
Interval |
E-value |
| A2M_2 |
cd02897 |
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy ... |
958-1264 |
8.17e-157 |
|
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy zone protein (PZP). Alpha(2)-M and PZP are broadly specific proteinase inhibitors. Alpha (2)-M is a major carrier protein in serum. The structural thioester of alpha (2)-M, is involved in the immobilization and entrapment of proteases. PZP is a trace protein in the plasma of non-pregnant females and males which is elevated in pregnancy. Alpha (2)-M and PZ bind to placental protein-14 and may modulate its activity in T-cell growth and cytokine production contributing to fetal survival. It has been suggested that thioester bond cleavage promotes the binding of PZ and alpha (2)-M to the CD91 receptor clearing them from circulation.
:
Pssm-ID: 239227 Cd Length: 292 Bit Score: 476.30 E-value: 8.17e-157
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 958 AMQNTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGERYGrnQG 1037
Cdd:cd02897 1 ALQNLDNLLRMPYGCGEQNMVNFAPNIYVLDYLKATGQLTPEIESKALGFLRTGYQRQLTYKHSDGSYSAFGESDK--SG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1038 NTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSAYITIALLEIPLTVT 1117
Cdd:cd02897 79 STWLTAFVLKSFAQARPFIYIDENVLQQALTWLSSHQKSNGCFREVGRVFHKAMQGGVDDEVALTAYVLIALLEAGLPSE 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1118 HPVVRNALFCLESAWKTAqegdhgSHVYTKALLAYAFALAGnQDKRKEVLKSLNEEAVKKDNSVHWERPqkPKAPVGHFY 1197
Cdd:cd02897 159 RPVVEKALSCLEAALDSI------SDPYTLALAAYALTLAG-SEKRPEALKKLDELAISEDGTKHWSRP--PPSEEGPSY 229
|
250 260 270 280 290 300
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 578822814 1198 EPQAPSAEVEMTSYVLLAYLTAQPaptsEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKY 1264
Cdd:cd02897 230 YWQAPSAEVEMTAYALLALLSAGG----EDLAEALPIVKWLAKQRNSLGGFSSTQDTVVALQALAKY 292
|
|
| A2M |
pfam00207 |
Alpha-2-macroglobulin family; This family includes the C-terminal region of the ... |
738-828 |
1.72e-41 |
|
Alpha-2-macroglobulin family; This family includes the C-terminal region of the alpha-2-macroglobulin family.
:
Pssm-ID: 459711 [Multi-domain] Cd Length: 91 Bit Score: 147.35 E-value: 1.72e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 738 TWIWDLVVVNSAGVAEVGVTVPDTITEWKAGAFCLSEDAGLGISSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNY 817
Cdd:pfam00207 1 TWLWDPVLVTDNGKASLSFTLPDSITTWRATAFALSPDTGLGVAEPPELVVFKPFFVDLNLPYSVRRGEQFELKATVFNY 80
|
90
....*....|.
gi 578822814 818 LPKCIRVSVQL 828
Cdd:pfam00207 81 LDKCLKVRVRL 91
|
|
| YfaS super family |
cl34462 |
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function ... |
129-1338 |
1.58e-38 |
|
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function prediction only];
The actual alignment was detected with superfamily member COG2373:
Pssm-ID: 441940 [Multi-domain] Cd Length: 1605 Bit Score: 157.94 E-value: 1.58e-38
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 129 VFVQTDKSIYKPGQTVKFRVVSMDENFHPLNELiPL-VYIQDPKGNRIAQwQSFQL-EGGLKQFSFPLSSEPFQGSYKVV 206
Cdd:COG2373 371 AFLFTDRGIYRPGETVHLKALLRDADGKAPAGL-PLtLELTDPDGKEVRR-QTLTLnEFGGYSFSFPLPEDAPTGTWRLE 448
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 207 VQ-KKSGGRTEHPFTVEEFVLPKFEVQVTVPKIITILEEEMNVSVCGLYTYGKPVPG-HVTVSICRK------------- 271
Cdd:COG2373 449 LYvDPKPALGSKSFRVEEFKPPRFKVDLTLDKEPLKPGDPVTVTVDARYLFGAPAAGlKVEGEVTLRpartafpgypgyr 528
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 272 YSDASDCHGEDSQAFcekFSGQLNSHGCFyqQVKTKVFQLKRKEYEMKLHTEAQIQEEGtvveltGRqssEITRTITkls 351
Cdd:COG2373 529 FGDPDEEFEPEELDL---GEGTLDADGKA--SLSLPLPDAPDAPGPLRATVEASVFESG------GR---PVTRSAT--- 591
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 352 fvkvdshfrqgIPFFGQVRLVdgkGVPIPNkvifirgneanyysNATTDEHGLVQFSINTTN-----VMGTSLTVRVNYK 426
Cdd:COG2373 592 -----------VPVHPADFYV---GIRLPL--------------FDGDPEGAPATFEVVAVDpdgkpVAGKGLKVELYRE 643
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 427 DRSpcY--------GYQWVSEEHEEAHHTAYLVFSPSK----SFVHLEPMSHEL----PCGHTQTVQAHYI-------LN 483
Cdd:COG2373 644 EWR--YvwyksddgGWRYESQEKEEPVAEGTLTTGADGpaslSLTPVEWGRYRLevkdPDGGLATSVRFYAggnaswgAE 721
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 484 GGTLLGLK------------KLSFY------YLIMakggIVRTGThgLLVKQEDMK-GHFSISIPVKSDIAPVARLLIYA 544
Cdd:COG2373 722 RPDRLELSldkesykpgetaKLLIQspfagrALVT----VERDGV--LETQWVDVKgGGTTVEIPVTEDWAPNAYVSATL 795
|
490 500 510 520 530 540 550 560
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 545 VLPTGDVIGDSAKY-------DVENcLANKVDLSFSPSQSL-PASHAHLRVTAA-----PQSVcALRAVDQSVLlmkpda 611
Cdd:COG2373 796 VRPGDSTANDMPARaygvaplPVDP-PARRLKVELTAPEKLrPGETLTVTVKVKgaagkAAEV-TLAAVDEGIL------ 867
|
570 580 590 600 610 620 630 640
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 612 elsassvynllpekDLTGF--PGPLNdqdnedcinrhnvYINGITYTPVSSTnekDMYSFLEDmglkaftnskirkpkmc 689
Cdd:COG2373 868 --------------NLTGYktPDPLD-------------FFYGKRALGVETR---DLYGRLIG----------------- 900
|
650 660 670 680 690 700 710 720
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 690 pqlqqyEMHGPEG-LRVGfyesdvmGRGHARLVHVEEPhtetVRKYFPETWIWD-LVVVNSAGVAEVGVTVPDTITEWKA 767
Cdd:COG2373 901 ------AFGGAAGaLRSG-------GDGALGRGGNPKP----PRKRFKPVALFSgPVKTDADGKATVSFDLPDFNGTLRV 963
|
730 740 750 760 770 780 790 800
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 768 GAFCLSEDAgLGiSSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNYLPKCIRVSVQLEASPAfLAVPVEKEQAPHc 847
Cdd:COG2373 964 MAVAWSDDR-FG-SAEATVTVRKPLVVRPSLPRFLAPGDRFELPVDVFNLTGKAGTVTVTLEASGG-LTLEGEATQTVT- 1039
|
810 820 830 840 850 860 870 880
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 848 ICANGRQTVSWAVTPKSLGNVNFTVSAEALesqelcgtevpsvpehGRKDTVIKPLLVEP-EGLEKETTFNSLlcpSGGE 926
Cdd:COG2373 1040 LAAGGRATVRFPLKAPDAGDAKVTVTATGG----------------GESDAREVELPVRPaNPLVTRATSGVL---APGE 1100
|
890 900 910 920 930 940 950 960
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 927 vSEELSLKLPPNVVEESARASVSV----LGDILGSAMQntqnLLQMPYGCGEQNMVLFAPNIYVLDyLNETQQLTPEIKS 1002
Cdd:COG2373 1101 -SWTLPLDLPGGLRPGTGSLTLSLssspPLDLAGLLRY----LLRYPYGCTEQTTSRALPLLYLSD-LAEALGLKGDKDA 1174
|
970 980 990 1000 1010 1020 1030 1040
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1003 KAIGYLNTGYQRQLNYKHYDGSYSTFGeryGRNQGNTWLTAFVLKTFAQARAY-IFIDEAHITQALIWLSQRQKDNGcfr 1081
Cdd:COG2373 1175 ELRARIQAAIARLLSMQNSDGGFGLWP---GGSESDPWLTAYATDFLLEAREAgYAVPDDALDRALDYLRNYLRNPW--- 1248
|
1050 1060 1070 1080 1090 1100 1110 1120
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1082 ssgsllnnAIKGGVEDEVTLSAYITIALleipltvthpvvrnALfclesawktAQEGDHGSHVYTKALLAYAF------- 1154
Cdd:COG2373 1249 --------EIEYDDAYRLAVRAYALYVL--------------AR---------AGKADLGDLRYLYDRRKDALsplakaq 1297
|
1130 1140 1150 1160 1170 1180 1190 1200
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1155 -----ALAGNQDKRKEVLKSLNEEAVKKDNSVHWerpqkpkapvGHFYepqapSAEVEMTSYVLLAYLTAQPAPtsedlT 1229
Cdd:COG2373 1298 laaalALLGDKARAEELLAAALARLRETGARDYW----------YGDY-----GSPLRDQALALALLAELGPDA-----P 1357
|
1210 1220 1230 1240 1250 1260 1270 1280
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1230 SATNIVKWITKQQNaQGGFSSTQDTVVALHALSKYGAATFTRTGKAAQVTIqsSGTfssKFQVDNNNRLLLQQVSLPEL- 1308
Cdd:COG2373 1358 LAPKLARWLAKALK-SGRWLSTQETAWALLALAAYARAAGASPDFTATLTL--DGK---TLPLTGRGPLARVTLPAAELl 1431
|
1290 1300 1310
....*....|....*....|....*....|
gi 578822814 1309 PGEYSMKVTGEGCVYLQTSLKYNILPEKEE 1338
Cdd:COG2373 1432 AGPLTITNTGDGPLYYTLTLSGYPAEGPPP 1461
|
|
| A2M_recep |
pfam07677 |
A-macroglobulin receptor binding domain; This family includes the receptor binding domain ... |
1374-1464 |
7.61e-34 |
|
A-macroglobulin receptor binding domain; This family includes the receptor binding domain region of the alpha-2-macroglobulin family.
:
Pssm-ID: 462226 Cd Length: 92 Bit Score: 125.37 E-value: 7.61e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1374 SASNMAIVDVKMVSGFIPLKPTVKMLERSNHVSRTE-VSSNHVLIYLDKVSNQTLSLFFTVLQDVPVRDLKPAIVKVYDY 1452
Cdd:pfam07677 1 ESSNMAILEVGLPSGFVPDEEDLKKLGVDPLIKRVEtVDDGKVILYLDKLSGEPLCFSFRAEQTFPVANLKPAPVKVYDY 80
|
90
....*....|..
gi 578822814 1453 YETDEFAIAEYN 1464
Cdd:pfam07677 81 YEPERRATTFYS 92
|
|
| |
|
Name |
Accession |
Description |
Interval |
E-value |
| A2M_2 |
cd02897 |
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy ... |
958-1264 |
8.17e-157 |
|
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy zone protein (PZP). Alpha(2)-M and PZP are broadly specific proteinase inhibitors. Alpha (2)-M is a major carrier protein in serum. The structural thioester of alpha (2)-M, is involved in the immobilization and entrapment of proteases. PZP is a trace protein in the plasma of non-pregnant females and males which is elevated in pregnancy. Alpha (2)-M and PZ bind to placental protein-14 and may modulate its activity in T-cell growth and cytokine production contributing to fetal survival. It has been suggested that thioester bond cleavage promotes the binding of PZ and alpha (2)-M to the CD91 receptor clearing them from circulation.
Pssm-ID: 239227 Cd Length: 292 Bit Score: 476.30 E-value: 8.17e-157
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 958 AMQNTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGERYGrnQG 1037
Cdd:cd02897 1 ALQNLDNLLRMPYGCGEQNMVNFAPNIYVLDYLKATGQLTPEIESKALGFLRTGYQRQLTYKHSDGSYSAFGESDK--SG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1038 NTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSAYITIALLEIPLTVT 1117
Cdd:cd02897 79 STWLTAFVLKSFAQARPFIYIDENVLQQALTWLSSHQKSNGCFREVGRVFHKAMQGGVDDEVALTAYVLIALLEAGLPSE 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1118 HPVVRNALFCLESAWKTAqegdhgSHVYTKALLAYAFALAGnQDKRKEVLKSLNEEAVKKDNSVHWERPqkPKAPVGHFY 1197
Cdd:cd02897 159 RPVVEKALSCLEAALDSI------SDPYTLALAAYALTLAG-SEKRPEALKKLDELAISEDGTKHWSRP--PPSEEGPSY 229
|
250 260 270 280 290 300
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 578822814 1198 EPQAPSAEVEMTSYVLLAYLTAQPaptsEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKY 1264
Cdd:cd02897 230 YWQAPSAEVEMTAYALLALLSAGG----EDLAEALPIVKWLAKQRNSLGGFSSTQDTVVALQALAKY 292
|
|
| TED_complement |
pfam07678 |
A-macroglobulin TED domain; This entry corresponds to the TED domain of the complement ... |
948-1264 |
1.95e-152 |
|
A-macroglobulin TED domain; This entry corresponds to the TED domain of the complement components such as C3, C4 and C5. This domain contains a short highly conserved region of proteinase-binding alpha-macro-globulins contains the cysteine and a glutamine of a thiol-ester bond that is cleaved at the moment of proteinase binding, and mediates the covalent binding of the alpha-macro-globulin to the proteinase. The GCGEQ motif is highly conserved.
Pssm-ID: 462227 [Multi-domain] Cd Length: 311 Bit Score: 465.62 E-value: 1.95e-152
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 948 VSVLGDILGSAMQ----NTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDG 1023
Cdd:pfam07678 1 ISVVGDIMGPAIQvvpeNLSSLLRLPYGCGEQNMVLFAPNVYVLRYLDKTNQLTKLIKSKAIDYLEQGYQRQLSYKHPDG 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1024 SYSTFGERygrnQGNTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSA 1103
Cdd:pfam07678 81 SYSAFGHS----PGSTWLTAFVLKVFAQARKFIFIDPEEICQSLRWLLSQQKPDGSFREPGPLLHRAMKGGVDGEVSLTA 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1104 YITIALLEI-----PLTVTHPVVRNALFCLESAwktaqEGDHGSHVYTKALLAYAFALAGNQDKRKEVLKSLNEEAVKKD 1178
Cdd:pfam07678 157 YVTIALLEAldingLLQRVHPSIRKALTYLEQA-----QLAGLTSPYTLAILAYALALAGSPETREELLKSLDAMAREEG 231
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1179 NSVHWERPQKPKAPVGHFYEPQAPSAEVEMTSYVLLAYLtaqpapTSEDLTSATNIVKWITKQQNAQGGFSSTQDTVVAL 1258
Cdd:pfam07678 232 NSRYWERDEKSDPQGVPEYPPQAPSLEVETTAYALLAYL------LLGDLTYADPIVKWLTSQRNSHGGFSSTQDTVVAL 305
|
....*.
gi 578822814 1259 HALSKY 1264
Cdd:pfam07678 306 QALAEY 311
|
|
| A2M |
pfam00207 |
Alpha-2-macroglobulin family; This family includes the C-terminal region of the ... |
738-828 |
1.72e-41 |
|
Alpha-2-macroglobulin family; This family includes the C-terminal region of the alpha-2-macroglobulin family.
Pssm-ID: 459711 [Multi-domain] Cd Length: 91 Bit Score: 147.35 E-value: 1.72e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 738 TWIWDLVVVNSAGVAEVGVTVPDTITEWKAGAFCLSEDAGLGISSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNY 817
Cdd:pfam00207 1 TWLWDPVLVTDNGKASLSFTLPDSITTWRATAFALSPDTGLGVAEPPELVVFKPFFVDLNLPYSVRRGEQFELKATVFNY 80
|
90
....*....|.
gi 578822814 818 LPKCIRVSVQL 828
Cdd:pfam00207 81 LDKCLKVRVRL 91
|
|
| YfaS |
COG2373 |
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function ... |
129-1338 |
1.58e-38 |
|
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function prediction only];
Pssm-ID: 441940 [Multi-domain] Cd Length: 1605 Bit Score: 157.94 E-value: 1.58e-38
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 129 VFVQTDKSIYKPGQTVKFRVVSMDENFHPLNELiPL-VYIQDPKGNRIAQwQSFQL-EGGLKQFSFPLSSEPFQGSYKVV 206
Cdd:COG2373 371 AFLFTDRGIYRPGETVHLKALLRDADGKAPAGL-PLtLELTDPDGKEVRR-QTLTLnEFGGYSFSFPLPEDAPTGTWRLE 448
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 207 VQ-KKSGGRTEHPFTVEEFVLPKFEVQVTVPKIITILEEEMNVSVCGLYTYGKPVPG-HVTVSICRK------------- 271
Cdd:COG2373 449 LYvDPKPALGSKSFRVEEFKPPRFKVDLTLDKEPLKPGDPVTVTVDARYLFGAPAAGlKVEGEVTLRpartafpgypgyr 528
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 272 YSDASDCHGEDSQAFcekFSGQLNSHGCFyqQVKTKVFQLKRKEYEMKLHTEAQIQEEGtvveltGRqssEITRTITkls 351
Cdd:COG2373 529 FGDPDEEFEPEELDL---GEGTLDADGKA--SLSLPLPDAPDAPGPLRATVEASVFESG------GR---PVTRSAT--- 591
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 352 fvkvdshfrqgIPFFGQVRLVdgkGVPIPNkvifirgneanyysNATTDEHGLVQFSINTTN-----VMGTSLTVRVNYK 426
Cdd:COG2373 592 -----------VPVHPADFYV---GIRLPL--------------FDGDPEGAPATFEVVAVDpdgkpVAGKGLKVELYRE 643
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 427 DRSpcY--------GYQWVSEEHEEAHHTAYLVFSPSK----SFVHLEPMSHEL----PCGHTQTVQAHYI-------LN 483
Cdd:COG2373 644 EWR--YvwyksddgGWRYESQEKEEPVAEGTLTTGADGpaslSLTPVEWGRYRLevkdPDGGLATSVRFYAggnaswgAE 721
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 484 GGTLLGLK------------KLSFY------YLIMakggIVRTGThgLLVKQEDMK-GHFSISIPVKSDIAPVARLLIYA 544
Cdd:COG2373 722 RPDRLELSldkesykpgetaKLLIQspfagrALVT----VERDGV--LETQWVDVKgGGTTVEIPVTEDWAPNAYVSATL 795
|
490 500 510 520 530 540 550 560
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 545 VLPTGDVIGDSAKY-------DVENcLANKVDLSFSPSQSL-PASHAHLRVTAA-----PQSVcALRAVDQSVLlmkpda 611
Cdd:COG2373 796 VRPGDSTANDMPARaygvaplPVDP-PARRLKVELTAPEKLrPGETLTVTVKVKgaagkAAEV-TLAAVDEGIL------ 867
|
570 580 590 600 610 620 630 640
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 612 elsassvynllpekDLTGF--PGPLNdqdnedcinrhnvYINGITYTPVSSTnekDMYSFLEDmglkaftnskirkpkmc 689
Cdd:COG2373 868 --------------NLTGYktPDPLD-------------FFYGKRALGVETR---DLYGRLIG----------------- 900
|
650 660 670 680 690 700 710 720
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 690 pqlqqyEMHGPEG-LRVGfyesdvmGRGHARLVHVEEPhtetVRKYFPETWIWD-LVVVNSAGVAEVGVTVPDTITEWKA 767
Cdd:COG2373 901 ------AFGGAAGaLRSG-------GDGALGRGGNPKP----PRKRFKPVALFSgPVKTDADGKATVSFDLPDFNGTLRV 963
|
730 740 750 760 770 780 790 800
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 768 GAFCLSEDAgLGiSSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNYLPKCIRVSVQLEASPAfLAVPVEKEQAPHc 847
Cdd:COG2373 964 MAVAWSDDR-FG-SAEATVTVRKPLVVRPSLPRFLAPGDRFELPVDVFNLTGKAGTVTVTLEASGG-LTLEGEATQTVT- 1039
|
810 820 830 840 850 860 870 880
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 848 ICANGRQTVSWAVTPKSLGNVNFTVSAEALesqelcgtevpsvpehGRKDTVIKPLLVEP-EGLEKETTFNSLlcpSGGE 926
Cdd:COG2373 1040 LAAGGRATVRFPLKAPDAGDAKVTVTATGG----------------GESDAREVELPVRPaNPLVTRATSGVL---APGE 1100
|
890 900 910 920 930 940 950 960
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 927 vSEELSLKLPPNVVEESARASVSV----LGDILGSAMQntqnLLQMPYGCGEQNMVLFAPNIYVLDyLNETQQLTPEIKS 1002
Cdd:COG2373 1101 -SWTLPLDLPGGLRPGTGSLTLSLssspPLDLAGLLRY----LLRYPYGCTEQTTSRALPLLYLSD-LAEALGLKGDKDA 1174
|
970 980 990 1000 1010 1020 1030 1040
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1003 KAIGYLNTGYQRQLNYKHYDGSYSTFGeryGRNQGNTWLTAFVLKTFAQARAY-IFIDEAHITQALIWLSQRQKDNGcfr 1081
Cdd:COG2373 1175 ELRARIQAAIARLLSMQNSDGGFGLWP---GGSESDPWLTAYATDFLLEAREAgYAVPDDALDRALDYLRNYLRNPW--- 1248
|
1050 1060 1070 1080 1090 1100 1110 1120
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1082 ssgsllnnAIKGGVEDEVTLSAYITIALleipltvthpvvrnALfclesawktAQEGDHGSHVYTKALLAYAF------- 1154
Cdd:COG2373 1249 --------EIEYDDAYRLAVRAYALYVL--------------AR---------AGKADLGDLRYLYDRRKDALsplakaq 1297
|
1130 1140 1150 1160 1170 1180 1190 1200
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1155 -----ALAGNQDKRKEVLKSLNEEAVKKDNSVHWerpqkpkapvGHFYepqapSAEVEMTSYVLLAYLTAQPAPtsedlT 1229
Cdd:COG2373 1298 laaalALLGDKARAEELLAAALARLRETGARDYW----------YGDY-----GSPLRDQALALALLAELGPDA-----P 1357
|
1210 1220 1230 1240 1250 1260 1270 1280
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1230 SATNIVKWITKQQNaQGGFSSTQDTVVALHALSKYGAATFTRTGKAAQVTIqsSGTfssKFQVDNNNRLLLQQVSLPEL- 1308
Cdd:COG2373 1358 LAPKLARWLAKALK-SGRWLSTQETAWALLALAAYARAAGASPDFTATLTL--DGK---TLPLTGRGPLARVTLPAAELl 1431
|
1290 1300 1310
....*....|....*....|....*....|
gi 578822814 1309 PGEYSMKVTGEGCVYLQTSLKYNILPEKEE 1338
Cdd:COG2373 1432 AGPLTITNTGDGPLYYTLTLSGYPAEGPPP 1461
|
|
| MG4 |
pfam17789 |
Macroglobulin domain MG4; This domain is MG4 found in complement C3 and C5 proteins. |
350-445 |
2.07e-35 |
|
Macroglobulin domain MG4; This domain is MG4 found in complement C3 and C5 proteins.
Pssm-ID: 465507 Cd Length: 95 Bit Score: 130.06 E-value: 2.07e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 350 LSFVKVDSHFRQGIPFFGQVRLVDGKGVPIPNKVIFIRGNEANYYSNATTDEHGLVQFSINTTNvMGTSLTVRVNYKDRS 429
Cdd:pfam17789 1 ITFEKTPKYFKPGLPFSGQVLVVDPDGSPAPNVPVFIEAGNTEFNQNLTTDEDGTAQFSINTPG-NAASLSITVKTKDPD 79
|
90
....*....|....*.
gi 578822814 430 PCYGYQWVSEEHEEAH 445
Cdd:pfam17789 80 LCPEHQALAEMYAEAY 95
|
|
| A2M_recep |
pfam07677 |
A-macroglobulin receptor binding domain; This family includes the receptor binding domain ... |
1374-1464 |
7.61e-34 |
|
A-macroglobulin receptor binding domain; This family includes the receptor binding domain region of the alpha-2-macroglobulin family.
Pssm-ID: 462226 Cd Length: 92 Bit Score: 125.37 E-value: 7.61e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1374 SASNMAIVDVKMVSGFIPLKPTVKMLERSNHVSRTE-VSSNHVLIYLDKVSNQTLSLFFTVLQDVPVRDLKPAIVKVYDY 1452
Cdd:pfam07677 1 ESSNMAILEVGLPSGFVPDEEDLKKLGVDPLIKRVEtVDDGKVILYLDKLSGEPLCFSFRAEQTFPVANLKPAPVKVYDY 80
|
90
....*....|..
gi 578822814 1453 YETDEFAIAEYN 1464
Cdd:pfam07677 81 YEPERRATTFYS 92
|
|
| |
|
Name |
Accession |
Description |
Interval |
E-value |
| A2M_2 |
cd02897 |
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy ... |
958-1264 |
8.17e-157 |
|
Proteins similar to alpha2-macroglobulin (alpha (2)-M). This group also contains the pregnancy zone protein (PZP). Alpha(2)-M and PZP are broadly specific proteinase inhibitors. Alpha (2)-M is a major carrier protein in serum. The structural thioester of alpha (2)-M, is involved in the immobilization and entrapment of proteases. PZP is a trace protein in the plasma of non-pregnant females and males which is elevated in pregnancy. Alpha (2)-M and PZ bind to placental protein-14 and may modulate its activity in T-cell growth and cytokine production contributing to fetal survival. It has been suggested that thioester bond cleavage promotes the binding of PZ and alpha (2)-M to the CD91 receptor clearing them from circulation.
Pssm-ID: 239227 Cd Length: 292 Bit Score: 476.30 E-value: 8.17e-157
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 958 AMQNTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGERYGrnQG 1037
Cdd:cd02897 1 ALQNLDNLLRMPYGCGEQNMVNFAPNIYVLDYLKATGQLTPEIESKALGFLRTGYQRQLTYKHSDGSYSAFGESDK--SG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1038 NTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSAYITIALLEIPLTVT 1117
Cdd:cd02897 79 STWLTAFVLKSFAQARPFIYIDENVLQQALTWLSSHQKSNGCFREVGRVFHKAMQGGVDDEVALTAYVLIALLEAGLPSE 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1118 HPVVRNALFCLESAWKTAqegdhgSHVYTKALLAYAFALAGnQDKRKEVLKSLNEEAVKKDNSVHWERPqkPKAPVGHFY 1197
Cdd:cd02897 159 RPVVEKALSCLEAALDSI------SDPYTLALAAYALTLAG-SEKRPEALKKLDELAISEDGTKHWSRP--PPSEEGPSY 229
|
250 260 270 280 290 300
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 578822814 1198 EPQAPSAEVEMTSYVLLAYLTAQPaptsEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKY 1264
Cdd:cd02897 230 YWQAPSAEVEMTAYALLALLSAGG----EDLAEALPIVKWLAKQRNSLGGFSSTQDTVVALQALAKY 292
|
|
| TED_complement |
pfam07678 |
A-macroglobulin TED domain; This entry corresponds to the TED domain of the complement ... |
948-1264 |
1.95e-152 |
|
A-macroglobulin TED domain; This entry corresponds to the TED domain of the complement components such as C3, C4 and C5. This domain contains a short highly conserved region of proteinase-binding alpha-macro-globulins contains the cysteine and a glutamine of a thiol-ester bond that is cleaved at the moment of proteinase binding, and mediates the covalent binding of the alpha-macro-globulin to the proteinase. The GCGEQ motif is highly conserved.
Pssm-ID: 462227 [Multi-domain] Cd Length: 311 Bit Score: 465.62 E-value: 1.95e-152
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 948 VSVLGDILGSAMQ----NTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDG 1023
Cdd:pfam07678 1 ISVVGDIMGPAIQvvpeNLSSLLRLPYGCGEQNMVLFAPNVYVLRYLDKTNQLTKLIKSKAIDYLEQGYQRQLSYKHPDG 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1024 SYSTFGERygrnQGNTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSA 1103
Cdd:pfam07678 81 SYSAFGHS----PGSTWLTAFVLKVFAQARKFIFIDPEEICQSLRWLLSQQKPDGSFREPGPLLHRAMKGGVDGEVSLTA 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1104 YITIALLEI-----PLTVTHPVVRNALFCLESAwktaqEGDHGSHVYTKALLAYAFALAGNQDKRKEVLKSLNEEAVKKD 1178
Cdd:pfam07678 157 YVTIALLEAldingLLQRVHPSIRKALTYLEQA-----QLAGLTSPYTLAILAYALALAGSPETREELLKSLDAMAREEG 231
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1179 NSVHWERPQKPKAPVGHFYEPQAPSAEVEMTSYVLLAYLtaqpapTSEDLTSATNIVKWITKQQNAQGGFSSTQDTVVAL 1258
Cdd:pfam07678 232 NSRYWERDEKSDPQGVPEYPPQAPSLEVETTAYALLAYL------LLGDLTYADPIVKWLTSQRNSHGGFSSTQDTVVAL 305
|
....*.
gi 578822814 1259 HALSKY 1264
Cdd:pfam07678 306 QALAEY 311
|
|
| A2M_like |
cd02891 |
Proteins similar to alpha2-macroglobulin (alpha (2)-M). Alpha (2)-M is a major carrier ... |
958-1264 |
9.22e-107 |
|
Proteins similar to alpha2-macroglobulin (alpha (2)-M). Alpha (2)-M is a major carrier protein in serum. It is a broadly specific proteinase inhibitor. The structural thioester of alpha (2)-M, is involved in the immobilization and entrapment of proteases. This group contains another broadly specific proteinase inhibitor: pregnancy zone protein (PZP). PZP is a trace protein in the plasma of non-pregnant females and males which is elevated in pregnancy. Alpha (2)-M and PZ bind to placental protein-14 and may modulate its activity in T-cell growth and cytokine production thereby protecting the allogeneic fetus from attack by the maternal immune system. This group also contains C3, C4 and C5 of vertebrate complement. The vertebrate complement is an effector of both the acquired and innate immune systems The point of convergence of the classical, alternative and lectin pathways of the complement system is the proteolytic activation of C3. C4 plays a key role in propagating the classical and lectin pathways. C5 participates in the classical and alternative pathways. The thioester bond located within the structure of C3 and C4 is central to the function of complement. C5 does not contain an active thioester bond.
Pssm-ID: 239221 [Multi-domain] Cd Length: 282 Bit Score: 340.52 E-value: 9.22e-107
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 958 AMQNTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQLTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGeryGRNQG 1037
Cdd:cd02891 1 SLGNLDYLLRYPYGCGEQTMSRAAPNLYVLKYLDATGQLTPEIREKALEYIRKGYQRLLTYQRSDGSFSAWG---NSDSG 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1038 NTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGVEDEVTLSAYITIALLEIPLTVt 1117
Cdd:cd02891 78 STWLTAYVVKFLSQARKYIDVDENVLARALGWLVPQQKEDGSFRELGPVIHREMKGGVDDSVSLTAYVLIALAEAGKAC- 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1118 HPVVRNALFCLESAWktaqegDHGSHVYTKALLAYAFALAGNQDKRKEVLKSLNEEAVKKDNSVHWERpqkpkapvgHFY 1197
Cdd:cd02891 157 DASIEKALAYLETQL------DGLLDPYALAILAYALALAGDSTRADEALKKLLEAAREKGGTAHWSL---------SWP 221
|
250 260 270 280 290 300
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 578822814 1198 EPQAPSAEVEMTSYVLLAYLTAqpaptsEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKY 1264
Cdd:cd02891 222 GDYGSSLRVEATAYALLALLKL------GDLEEAGPIAKWLAQQRNSGGGFLSTQDTVVALQALAAY 282
|
|
| complement_C3_C4_C5 |
cd02896 |
Proteins similar to C3, C4 and C5 of vertebrate complement. The vertebrate complement system, ... |
961-1264 |
4.51e-71 |
|
Proteins similar to C3, C4 and C5 of vertebrate complement. The vertebrate complement system, comprised of a large number of distinct plasma proteins, is an effector of both the acquired and innate immune systems. The point of convergence of the classical, alternative and lectin pathways of the complement system is the proteolytic activation of C3. C4 plays a key role in propagating the classical and lectin pathways. C5 participates in the classical and alternative pathways. The thioester bond located within the structure of C3 and C4 is central to the function of complement. C5 does not contain an active thioester bond.
Pssm-ID: 239226 [Multi-domain] Cd Length: 297 Bit Score: 240.25 E-value: 4.51e-71
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 961 NTQNLLQMPYGCGEQNMVLFAPNIYVLDYLNETQQ---LTPEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFGERygrnQG 1037
Cdd:cd02896 4 GLEKLIRLPTGCGEQTMIKLAPTVYALRYLDTTNQwekLGPERRDEALKYIRQGYQRQLSYRKPDGSYAAWKNR----PS 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1038 NTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNAIKGGV---EDEVTLSAYITIALLEIpL 1114
Cdd:cd02896 80 STWLTAFVVKVFSLARKYIPVDQNVICGSVNWLISNQKPDGSFQEPSPVIHREMTGGVegsEGDVSLTAFVLIALQEA-R 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1115 TVTHPVVRNalfcLESAWKTAQE-----GDHGSHVYTKALLAYAFALAgNQDKRKEVLKSLNEEAVKKDNSVHWERPQKP 1189
Cdd:cd02896 159 SICPPEVQN----LDQSIRKAISylenqLPNLQRPYALAITAYALALA-DSPLSHAANRKLLSLAKRDGNGWYWWTIDSP 233
|
250 260 270 280 290 300 310
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 578822814 1190 kapvgHFYEPQAPSAEVEMTSYVLLAYLtaqpapTSEDLTSATNIVKWITKQQNAQGGFSSTQDTVVALHALSKY 1264
Cdd:cd02896 234 -----YWPVPGPSAITVETTAYALLALL------KLGDIEYANPIARWLTEQRNYGGGFGSTQDTVVALQALAEY 297
|
|
| ISOPREN_C2_like |
cd00688 |
This group contains class II terpene cyclases, protein prenyltransferases beta subunit, two ... |
958-1264 |
6.91e-61 |
|
This group contains class II terpene cyclases, protein prenyltransferases beta subunit, two broadly specific proteinase inhibitors alpha2-macroglobulin (alpha (2)-M) and pregnancy zone protein (PZP) and, the C3 C4 and C5 components of vertebrate complement. Class II terpene cyclases include squalene cyclase (SQCY) and 2,3-oxidosqualene cyclase (OSQCY), these integral membrane proteins catalyze a cationic cyclization cascade converting linear triterpenes to fused ring compounds. The protein prenyltransferases include protein farnesyltransferase (FTase) and geranylgeranyltransferase types I and II (GGTase-I and GGTase-II) which catalyze the carboxyl-terminal lipidation of Ras, Rab, and several other cellular signal transduction proteins, facilitating membrane associations and specific protein-protein interactions. Alpha (2)-M is a major carrier protein in serum and involved in the immobilization and entrapment of proteases. PZP is a pregnancy associated protein. Alpha (2)-M and PZP are known to bind to and, may modulate, the activity of placental protein-14 in T-cell growth and cytokine production thereby protecting the allogeneic fetus from attack by the maternal immune system.
Pssm-ID: 238362 [Multi-domain] Cd Length: 300 Bit Score: 211.26 E-value: 6.91e-61
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 958 AMQNTQNLLQMPYG--------CGEQNMVLFAPNIYVLDYLNETqqltpEIKSKAIGYLNTGYQRQLNYKHYDGSYSTFG 1029
Cdd:cd00688 1 IEKHLKYLLRYPYGdghwyqslCGEQTWSTAWPLLALLLLLAAT-----GIRDKADENIEKGIQRLLSYQLSDGGFSGWG 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1030 eryGRNQGNTWLTAFVLKTFAQARAYIFIDEAHITQALIWLSQRQKDNGCFRSSGSLLNNaiKGGVEDEVTLSAYITIAL 1109
Cdd:cd00688 76 ---GNDYPSLWLTAYALKALLLAGDYIAVDRIDLARALNWLLSLQNEDGGFREDGPGNHR--IGGDESDVRLTAYALIAL 150
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1110 LEIPLTVTHPVVRNALFCLESawktAQEGDHG------SHVYTKALLAYAFALAG--NQDKRKEVLKSLNEEAVKKDNSV 1181
Cdd:cd00688 151 ALLGKLDPDPLIEKALDYLLS----CQNYDGGfgpggeSHGYGTACAAAALALLGdlDSPDAKKALRWLLSRQRPDGGWG 226
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1182 HWERPqkpkapvghfYEPQAPSAEVEMTSYVLLAYLTAqpaptsEDLTSATNIVKWITKQQNAQGGFSS-------TQDT 1254
Cdd:cd00688 227 EGRDR----------TNKLSDSCYTEWAAYALLALGKL------GDLEDAEKLVKWLLSQQNEDGGFSSkpgksydTQHT 290
|
330
....*....|
gi 578822814 1255 VVALHALSKY 1264
Cdd:cd00688 291 VFALLALSLY 300
|
|
| A2M |
pfam00207 |
Alpha-2-macroglobulin family; This family includes the C-terminal region of the ... |
738-828 |
1.72e-41 |
|
Alpha-2-macroglobulin family; This family includes the C-terminal region of the alpha-2-macroglobulin family.
Pssm-ID: 459711 [Multi-domain] Cd Length: 91 Bit Score: 147.35 E-value: 1.72e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 738 TWIWDLVVVNSAGVAEVGVTVPDTITEWKAGAFCLSEDAGLGISSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNY 817
Cdd:pfam00207 1 TWLWDPVLVTDNGKASLSFTLPDSITTWRATAFALSPDTGLGVAEPPELVVFKPFFVDLNLPYSVRRGEQFELKATVFNY 80
|
90
....*....|.
gi 578822814 818 LPKCIRVSVQL 828
Cdd:pfam00207 81 LDKCLKVRVRL 91
|
|
| YfaS |
COG2373 |
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function ... |
129-1338 |
1.58e-38 |
|
Uncharacterized conserved protein YfaS, alpha-2-macroglobulin family [General function prediction only];
Pssm-ID: 441940 [Multi-domain] Cd Length: 1605 Bit Score: 157.94 E-value: 1.58e-38
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 129 VFVQTDKSIYKPGQTVKFRVVSMDENFHPLNELiPL-VYIQDPKGNRIAQwQSFQL-EGGLKQFSFPLSSEPFQGSYKVV 206
Cdd:COG2373 371 AFLFTDRGIYRPGETVHLKALLRDADGKAPAGL-PLtLELTDPDGKEVRR-QTLTLnEFGGYSFSFPLPEDAPTGTWRLE 448
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 207 VQ-KKSGGRTEHPFTVEEFVLPKFEVQVTVPKIITILEEEMNVSVCGLYTYGKPVPG-HVTVSICRK------------- 271
Cdd:COG2373 449 LYvDPKPALGSKSFRVEEFKPPRFKVDLTLDKEPLKPGDPVTVTVDARYLFGAPAAGlKVEGEVTLRpartafpgypgyr 528
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 272 YSDASDCHGEDSQAFcekFSGQLNSHGCFyqQVKTKVFQLKRKEYEMKLHTEAQIQEEGtvveltGRqssEITRTITkls 351
Cdd:COG2373 529 FGDPDEEFEPEELDL---GEGTLDADGKA--SLSLPLPDAPDAPGPLRATVEASVFESG------GR---PVTRSAT--- 591
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 352 fvkvdshfrqgIPFFGQVRLVdgkGVPIPNkvifirgneanyysNATTDEHGLVQFSINTTN-----VMGTSLTVRVNYK 426
Cdd:COG2373 592 -----------VPVHPADFYV---GIRLPL--------------FDGDPEGAPATFEVVAVDpdgkpVAGKGLKVELYRE 643
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 427 DRSpcY--------GYQWVSEEHEEAHHTAYLVFSPSK----SFVHLEPMSHEL----PCGHTQTVQAHYI-------LN 483
Cdd:COG2373 644 EWR--YvwyksddgGWRYESQEKEEPVAEGTLTTGADGpaslSLTPVEWGRYRLevkdPDGGLATSVRFYAggnaswgAE 721
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 484 GGTLLGLK------------KLSFY------YLIMakggIVRTGThgLLVKQEDMK-GHFSISIPVKSDIAPVARLLIYA 544
Cdd:COG2373 722 RPDRLELSldkesykpgetaKLLIQspfagrALVT----VERDGV--LETQWVDVKgGGTTVEIPVTEDWAPNAYVSATL 795
|
490 500 510 520 530 540 550 560
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 545 VLPTGDVIGDSAKY-------DVENcLANKVDLSFSPSQSL-PASHAHLRVTAA-----PQSVcALRAVDQSVLlmkpda 611
Cdd:COG2373 796 VRPGDSTANDMPARaygvaplPVDP-PARRLKVELTAPEKLrPGETLTVTVKVKgaagkAAEV-TLAAVDEGIL------ 867
|
570 580 590 600 610 620 630 640
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 612 elsassvynllpekDLTGF--PGPLNdqdnedcinrhnvYINGITYTPVSSTnekDMYSFLEDmglkaftnskirkpkmc 689
Cdd:COG2373 868 --------------NLTGYktPDPLD-------------FFYGKRALGVETR---DLYGRLIG----------------- 900
|
650 660 670 680 690 700 710 720
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 690 pqlqqyEMHGPEG-LRVGfyesdvmGRGHARLVHVEEPhtetVRKYFPETWIWD-LVVVNSAGVAEVGVTVPDTITEWKA 767
Cdd:COG2373 901 ------AFGGAAGaLRSG-------GDGALGRGGNPKP----PRKRFKPVALFSgPVKTDADGKATVSFDLPDFNGTLRV 963
|
730 740 750 760 770 780 790 800
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 768 GAFCLSEDAgLGiSSTASLRAFQPFFVELTMPYSVIRGEAFTLKATVLNYLPKCIRVSVQLEASPAfLAVPVEKEQAPHc 847
Cdd:COG2373 964 MAVAWSDDR-FG-SAEATVTVRKPLVVRPSLPRFLAPGDRFELPVDVFNLTGKAGTVTVTLEASGG-LTLEGEATQTVT- 1039
|
810 820 830 840 850 860 870 880
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 848 ICANGRQTVSWAVTPKSLGNVNFTVSAEALesqelcgtevpsvpehGRKDTVIKPLLVEP-EGLEKETTFNSLlcpSGGE 926
Cdd:COG2373 1040 LAAGGRATVRFPLKAPDAGDAKVTVTATGG----------------GESDAREVELPVRPaNPLVTRATSGVL---APGE 1100
|
890 900 910 920 930 940 950 960
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 927 vSEELSLKLPPNVVEESARASVSV----LGDILGSAMQntqnLLQMPYGCGEQNMVLFAPNIYVLDyLNETQQLTPEIKS 1002
Cdd:COG2373 1101 -SWTLPLDLPGGLRPGTGSLTLSLssspPLDLAGLLRY----LLRYPYGCTEQTTSRALPLLYLSD-LAEALGLKGDKDA 1174
|
970 980 990 1000 1010 1020 1030 1040
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1003 KAIGYLNTGYQRQLNYKHYDGSYSTFGeryGRNQGNTWLTAFVLKTFAQARAY-IFIDEAHITQALIWLSQRQKDNGcfr 1081
Cdd:COG2373 1175 ELRARIQAAIARLLSMQNSDGGFGLWP---GGSESDPWLTAYATDFLLEAREAgYAVPDDALDRALDYLRNYLRNPW--- 1248
|
1050 1060 1070 1080 1090 1100 1110 1120
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1082 ssgsllnnAIKGGVEDEVTLSAYITIALleipltvthpvvrnALfclesawktAQEGDHGSHVYTKALLAYAF------- 1154
Cdd:COG2373 1249 --------EIEYDDAYRLAVRAYALYVL--------------AR---------AGKADLGDLRYLYDRRKDALsplakaq 1297
|
1130 1140 1150 1160 1170 1180 1190 1200
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1155 -----ALAGNQDKRKEVLKSLNEEAVKKDNSVHWerpqkpkapvGHFYepqapSAEVEMTSYVLLAYLTAQPAPtsedlT 1229
Cdd:COG2373 1298 laaalALLGDKARAEELLAAALARLRETGARDYW----------YGDY-----GSPLRDQALALALLAELGPDA-----P 1357
|
1210 1220 1230 1240 1250 1260 1270 1280
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1230 SATNIVKWITKQQNaQGGFSSTQDTVVALHALSKYGAATFTRTGKAAQVTIqsSGTfssKFQVDNNNRLLLQQVSLPEL- 1308
Cdd:COG2373 1358 LAPKLARWLAKALK-SGRWLSTQETAWALLALAAYARAAGASPDFTATLTL--DGK---TLPLTGRGPLARVTLPAAELl 1431
|
1290 1300 1310
....*....|....*....|....*....|
gi 578822814 1309 PGEYSMKVTGEGCVYLQTSLKYNILPEKEE 1338
Cdd:COG2373 1432 AGPLTITNTGDGPLYYTLTLSGYPAEGPPP 1461
|
|
| MG4 |
pfam17789 |
Macroglobulin domain MG4; This domain is MG4 found in complement C3 and C5 proteins. |
350-445 |
2.07e-35 |
|
Macroglobulin domain MG4; This domain is MG4 found in complement C3 and C5 proteins.
Pssm-ID: 465507 Cd Length: 95 Bit Score: 130.06 E-value: 2.07e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 350 LSFVKVDSHFRQGIPFFGQVRLVDGKGVPIPNKVIFIRGNEANYYSNATTDEHGLVQFSINTTNvMGTSLTVRVNYKDRS 429
Cdd:pfam17789 1 ITFEKTPKYFKPGLPFSGQVLVVDPDGSPAPNVPVFIEAGNTEFNQNLTTDEDGTAQFSINTPG-NAASLSITVKTKDPD 79
|
90
....*....|....*.
gi 578822814 430 PCYGYQWVSEEHEEAH 445
Cdd:pfam17789 80 LCPEHQALAEMYAEAY 95
|
|
| A2M_recep |
pfam07677 |
A-macroglobulin receptor binding domain; This family includes the receptor binding domain ... |
1374-1464 |
7.61e-34 |
|
A-macroglobulin receptor binding domain; This family includes the receptor binding domain region of the alpha-2-macroglobulin family.
Pssm-ID: 462226 Cd Length: 92 Bit Score: 125.37 E-value: 7.61e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 1374 SASNMAIVDVKMVSGFIPLKPTVKMLERSNHVSRTE-VSSNHVLIYLDKVSNQTLSLFFTVLQDVPVRDLKPAIVKVYDY 1452
Cdd:pfam07677 1 ESSNMAILEVGLPSGFVPDEEDLKKLGVDPLIKRVEtVDDGKVILYLDKLSGEPLCFSFRAEQTFPVANLKPAPVKVYDY 80
|
90
....*....|..
gi 578822814 1453 YETDEFAIAEYN 1464
Cdd:pfam07677 81 YEPERRATTFYS 92
|
|
| A2M_BRD |
pfam07703 |
Alpha-2-macroglobulin bait region domain; Alpha-2-macroglobulins (A2Ms) are plasma proteins ... |
459-607 |
8.13e-34 |
|
Alpha-2-macroglobulin bait region domain; Alpha-2-macroglobulins (A2Ms) are plasma proteins that trap and inhibit a broad range of proteases and are major components of the eukaryotic innate immune system. However, A2M-like proteins were identified in pathogenically invasive bacteria and species that colonize higher eukaryotes. This domain is found in eukaryotic and bacterial proteins. In human A2Ms, this domain encompasses macroglobulin-like domain MG5 and 6 including bait region. In Salmonella enterica ser A2Ms, this domain encompasses MG7 and MG8 including the bait region. The Bait region is cleaved by proteases, followed by a large conformational change that blocks the target protease within a cage-like complex. This model of protease entrapment is recognized as the Venus flytrap mechanism.
Pssm-ID: 462235 [Multi-domain] Cd Length: 139 Bit Score: 127.08 E-value: 8.13e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 459 VHLEPMSHELPCGHTQTVQAHYILNGGTLLGlkklSFYYLIMAKGGIVRTGTHGllvkqedmkGHFSISIPVKSDIAPVA 538
Cdd:pfam07703 1 LHLSTDKTEYKPGETATVTVKSPFDGTVERD----GFTYLVLSKGQIVVVGRGG---------VTTSFSLPVTAEMAPSA 67
|
90 100 110 120 130 140 150
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 578822814 539 RLLIYAV---LPTGDVIGDSAKYDVENCLANKVDLSFSPSQSLPASHAHLRVTAAPQSVCALRAVDQSVLLM 607
Cdd:pfam07703 68 RVVAYYVrvdLSKPEVVADSVWVDVDDTCENKLKVTLSAEKYRPGSTVELKVKADPGAYVALAAVDKGVLLL 139
|
|
| MG3 |
pfam17791 |
Macroglobulin domain MG3; This entry corresponds to the MG3 domain found in complement ... |
223-313 |
9.03e-31 |
|
Macroglobulin domain MG3; This entry corresponds to the MG3 domain found in complement components C3, C4 and C5.
Pssm-ID: 465509 Cd Length: 83 Bit Score: 116.22 E-value: 9.03e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 223 EFVLPKFEVQVTVPKIITILEEEMNVSVCGLYTYGKPVPGHVTVSICRKYSDASDCHgedsqafcEKFSGQLNSHGCFYQ 302
Cdd:pfam17791 1 EYVLPKFEVKVEVPKFISVKDEEFQVTICAKYTYGKPVKGKAYVTLCLKDDSKRKCF--------ESFSKELDKDGCGSA 72
|
90
....*....|.
gi 578822814 303 QVKTKVFQLKR 313
Cdd:pfam17791 73 SLSTEEFQLTF 83
|
|
| MG2 |
pfam01835 |
MG2 domain; This is the MG2 (macroglobulin) domain of alpha-2-macroglobulin in eukaryotes. ... |
129-221 |
2.77e-25 |
|
MG2 domain; This is the MG2 (macroglobulin) domain of alpha-2-macroglobulin in eukaryotes. Alpha-2-macroglobulins (A2Ms) are plasma proteins that trap and inhibit a broad range of proteases and are major components of the eukaryotic innate immune system. However, A2M-like proteins were identified in pathogenically invasive bacteria and species that colonize higher eukaryotes. This domain is found in eukaryotic and bacterial proteins. In human A2Ms, this domain is termed macroglobulin-like (MG) domain 2 and in Salmonella enterica ser A2Ms, this is domain 4.
Pssm-ID: 426464 [Multi-domain] Cd Length: 95 Bit Score: 101.24 E-value: 2.77e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 578822814 129 VFVQTDKSIYKPGQTVKFRVVSMDENFHPLNELIPLVYIQDPKGNRIAQWQSFQLEGGLKQFSFPLSSEPFQGSYKVVVQ 208
Cdd:pfam01835 2 AFVYTDRGIYRPGETVHFKGLLRDQDLRPLAGLPVTLTVTDPDGNEVRRLPLTTDEFGGFSGSFPLPETAPTGTYTVVLR 81
|
90
....*....|....
gi 578822814 209 K-KSGGRTEHPFTV 221
Cdd:pfam01835 82 DgAGGSLGSGSFRV 95
|
|
| Big_1 |
pfam02369 |
Bacterial Ig-like domain (group 1); This family consists of bacterial domains with an Ig-like ... |
368-423 |
2.71e-03 |
|
Bacterial Ig-like domain (group 1); This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial surface proteins such as intimins and invasins involved in pathogenicity.
Pssm-ID: 460541 [Multi-domain] Cd Length: 64 Bit Score: 37.53 E-value: 2.71e-03
10 20 30 40 50
....*....|....*....|....*....|....*....|....*....|....*..
gi 578822814 368 QVRLVDGKGVPIPNK-VIFIRGNEANYYSNATTDEHGLVQFSInTTNVMGTSlTVRV 423
Cdd:pfam02369 10 TATVTDANGNPVPGAtVTFSASGGTLSASSGTTDANGQATVTL-TSTKAGTV-TVTA 64
|
|
| |