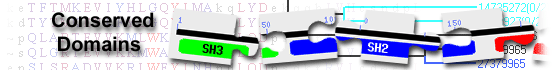|
Name |
Accession |
Description |
Interval |
E-value |
| rim_protein |
TIGR01257 |
retinal-specific rim ABC transporter; This model describes the photoreceptor protein (rim ... |
6-2238 |
0e+00 |
|
retinal-specific rim ABC transporter; This model describes the photoreceptor protein (rim protein) in eukaryotes. It is the member of ABC transporter superfamily. Rim protein is a membrane glycoprotein which is localized in the photoreceptor outer segment discs. Mutation/s in its genetic loci is implicated in the recessive Stargardt's disease. [Transport and binding proteins, Other]
Pssm-ID: 130324 [Multi-domain] Cd Length: 2272 Bit Score: 2468.68 E-value: 0e+00
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 6 QLRLLLWKNLTFRRRQTCQLLLEVAWPLFIFLILISVRLSYPPYEQHECHFPNKAMPSAGTLPWVQGIICNANNPCFRYP 85
Cdd:TIGR01257 6 QIQLLLWKNWTLRKRQKIRFVVELVWPLSLFLVLIWLRNANPLYSQHECHFPNKAMPSAGMLPWLQGIFCNVNNPCFQSP 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 86 TPGEAPGVVGNFNKSIVSRLFSDAQRLLLYSQRDTSIKDMHKVLRMLRQI-----KHPNS----NLKLQDFLVDNETFSG 156
Cdd:TIGR01257 86 TPGESPGIVSNYNNSILARVYRDFQELLMDAPESQHLGQVWAELRTLSQFmdtlrTHPERiagrGIRIRDILKDEEALTL 165
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 157 FLQHNLSLPRSTVDSLLQANVGLQKvFLQGY-QLHLASL-CNGSKLEEII---QLGDAEV--SALCGLPRKKLDAAERVL 229
Cdd:TIGR01257 166 FLMKNIGLSDSVVYLLVNSQVRPEQ-FAYGVpDLELKDIaCSEALLERFIifsQRRGAQTvrDALCSLSQGTLQWIEDTL 244
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 230 RYNMDILKPVVTklnstshLPTQHLAEATTVLLDSLGGL-------AQELFSTKSWSDMRQEVMFLTNVNSSSSSTQIYQ 302
Cdd:TIGR01257 245 YANVDFFKLFHV-------LPTLLDSRSQGINLRSWGGIlsdmsprIQEFIHRPSVQDLLWVTRPLLQNGGPETFTQLMG 317
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 303 AVSRIVCGHPEGGGLKIKSLNWYEDNNYKALFGGNNTEEDVDTFYDNSTTPYCNDLMKNLESSPLSRIIWKALKPLLVGK 382
Cdd:TIGR01257 318 ILSDLLCGYPEGGGSRVFSFNWYEDNNYKAFLGIDSTRKDPIYSYDKRTTSFCNALIQSLESNPLTKIAWRAAKPLLMGK 397
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 383 ILYTPDTPATRQVMAEVNKTFQELAVFHDLEGMWEELSPQIWTFMENSQEMDLVRTLLDSRGNDQFWEQKLDGLDWTAQD 462
Cdd:TIGR01257 398 ILFTPDSPAARRILKNANSTFEELERVRKLVKAWEEVGPQIWYFFDKSTQMTMIRDTLQNPTVKDFINRQLGEEGITAEA 477
|
490 500 510 520 530 540 550 560
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 463 IMAFLAKNPEDVQSPNGSVYTWREAFNETNQAIQTISRFMECVNLNKLEPIPTEVRLINKSMELLDERKFWAGIVFTGIT 542
Cdd:TIGR01257 478 VLNFLYNGPREKQADDMTNFDWRDIFNITDRFLRLANQYLECLVLDKFESYDDEVQLTQRALSLLEENRFWAGVVFPDMY 557
|
570 580 590 600 610 620 630 640
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 543 PDSVELPHHVKYKIRMDIDNVERTNKIKDGYWDPGPRADPFEDMRYVWGGFAYLQDVVEQAIIRVLTGSEKKTGVYVQQM 622
Cdd:TIGR01257 558 PWTSSLPPHVKYKIRMDIDVVEKTNKIKDRYWDSGPRADPVEDFRYIWGGFAYLQDMVEQGITRSQMQAEPPVGIYLQQM 637
|
650 660 670 680 690 700 710 720
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 623 PYPCYVDDIFLRVMSRSMPLFMTLAWIYSVAVIIKSIVYEKEARLKETMRIMGLDNGILWFSWFVSSLIPLLVSAGLLVV 702
Cdd:TIGR01257 638 PYPCFVDDSFMIILNRCFPIFMVLAWIYSVSMTVKSIVLEKELRLKETLKNQGVSNAVIWCTWFLDSFSIMSMSIFLLTI 717
|
730 740 750 760 770 780 790 800
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 703 ILKLGNLLPYSDPSVVFVFLSVFAMVTILQCFLISTLFSRANLAAACGGIIYFTLYLPYVLCVAWQDYVGFSIKIFASLL 782
Cdd:TIGR01257 718 FIMHGRILHYSDPFILFLFLLAFSTATIMQCFLLSTFFSKASLAAACSGVIYFTLYLPHILCFAWQDRMTADLKTAVSLL 797
|
810 820 830 840 850 860 870 880
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 783 SPVAFGFGCEYFALFEEQGIGVQWDNLFESPVEEDGFNLTTAVSMMLFDTFLYGVMTWYIEAVFPGQYGIPRPWYFPCTK 862
Cdd:TIGR01257 798 SPVAFGFGTEYLVRFEEQGLGLQWSNIGNSPLEGDEFSFLLSMKMMLLDAALYGLLAWYLDQVFPGDYGTPLPWYFLLQE 877
|
890 900 910 920 930 940 950 960
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 863 SYWFG-------------------EEIDEKSHPgssqKGVSEICMEEEPTHLRLGVSIQNLVKVYRDGMKVAVDGLALNF 923
Cdd:TIGR01257 878 SYWLGgegcstreeralektepltEEMEDPEHP----EGINDSFFERELPGLVPGVCVKNLVKIFEPSGRPAVDRLNITF 953
|
970 980 990 1000 1010 1020 1030 1040
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 924 YEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWFYARLKGL 1003
Cdd:TIGR01257 954 YENQITAFLGHNGAGKTTTLSILTGLLPPTSGTVLVGGKDIETNLDAVRQSLGMCPQHNILFHHLTVAEHILFYAQLKGR 1033
|
1050 1060 1070 1080 1090 1100 1110 1120
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1004 SEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRT 1083
Cdd:TIGR01257 1034 SWEEAQLEMEAMLEDTGL-HHKRNEEAQDLSGGMQRKLSVAIAFVGDAKVVVLDEPTSGVDPYSRRSIWDLLLKYRSGRT 1112
|
1130 1140 1150 1160 1170 1180 1190 1200
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1084 IILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQLGTGYYLTLVKKdVESSLSSCRNSSSTVSCLKKvvpGLpaAG 1163
Cdd:TIGR01257 1113 IIMSTHHMDEADLLGDRIAIISQGRLYCSGTPLFLKNCFGTGFYLTLVRK-MKNIQSQRGGCEGTCSCTSK---GF--ST 1186
|
1210 1220 1230 1240 1250 1260 1270 1280
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1164 QSGGRGGEVQRLVALTQDVSAISNLIRKHVSEARLVEDIGHELTYVLPYEAAKEGAFVELFHEIDDRLSDLGISSYGISE 1243
Cdd:TIGR01257 1187 RCPARVDEITPEQVLDGDVNELMDLVYHHVPEAKLVECIGQELIFLLPNKNFKQRAYASLFRELEETLADLGLSSFGISD 1266
|
1290 1300 1310 1320 1330 1340 1350 1360
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1244 TTLEEIFLKVAEESGVDAETSDGtlpARRNRRAFGDKQSCLHPfTEDDAVDPNDSDIDPESRETDLLSGM-----DGKGS 1318
Cdd:TIGR01257 1267 TPLEEIFLKVTEDADSGSLFAGG---AQQKRENANLRHPCSGP-TEKAGQTPQASHTCSPGQPAAHPEGQpppepEDPGV 1342
|
1370 1380 1390 1400 1410 1420 1430 1440
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1319 YQLKGWKLTQQQFVALLWKRLLIARRSRKGFFAQIVLPAVFVCIALVFSLIVPPFGKYPSLELQPWMYNEQYTFVSNDAP 1398
Cdd:TIGR01257 1343 PLNTGARLILQHVQALLVKRFQHTIRSHKDFLAQIVLPATFVFLALMLSIIIPPFGEYPALTLHPWMYGQQYTFFSMDEP 1422
|
1450 1460 1470 1480 1490 1500 1510 1520
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1399 EDMGTQELLNALTKDPGFGTRCMEGNPIPDTPClAGEEDWTISPVPQSIVDLFQNGNWTMKNPSPACQCSSDKIKKMLPV 1478
Cdd:TIGR01257 1423 NSEHLEVLADVLLNKPGFGNRCLKEEWLPEYPC-GNSTPWKTPSVSPNITHLFQKQKWTAAHPSPSCRCSTREKLTMLPE 1501
|
1530 1540 1550 1560 1570 1580 1590 1600
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1479 CPPGAGGLPPPQRKQKTADILQNLTGRNISDYLVKTYVQIIAKSLKNKIWVNEFRYGGFSLGvSNSQALPPSHE-VNDAI 1557
Cdd:TIGR01257 1502 CPEGAGGLPPPQRTQRSTEILQDLTDRNISDFLVKTYPALIRSSLKSKFWVNEQRYGGISIG-GKLPAIPITGEaLVGFL 1580
|
1610 1620 1630 1640 1650 1660 1670 1680
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1558 KQMKKLLKLTKDSSADRFLSSLGRFMAGLDTKNNVKVWFNNKGWHAISSFLNVINNAILRANLQKGENPSQYGITAFNHP 1637
Cdd:TIGR01257 1581 SDLGQMMNVSGGPVTREASKEMPDFLKHLETEDNIKVWFNNKGWHALVSFLNVAHNAILRASLPKDRDPEEYGITVISQP 1660
|
1690 1700 1710 1720 1730 1740 1750 1760
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1638 LNLTKQQLSEVALMTTSVDVLVSICVIFAMSFVPASFVVFLIQERVSKAKHLQFISGVKPVIYWLSNFVWDMCNYVVPAT 1717
Cdd:TIGR01257 1661 LNLTKEQLSEITVLTTSVDAVVAICVIFAMSFVPASFVLYLIQERVNKAKHLQFISGVSPTTYWLTNFLWDIMNYAVSAG 1740
|
1770 1780 1790 1800 1810 1820 1830 1840
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1718 LVIIIFICFQQKSYVSSTNLPVLALLLLLYGWSITPLMYPASFVFKIPSTAYVVLTSVNLFIGINGSVATFVLELFTNNK 1797
Cdd:TIGR01257 1741 LVVGIFIGFQKKAYTSPENLPALVALLMLYGWAVIPMMYPASFLFDVPSTAYVALSCANLFIGINSSAITFVLELFENNR 1820
|
1850 1860 1870 1880 1890 1900 1910 1920
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1798 -LNDINDILKSVFLIFPHFCLGRGLIDMVKNQAMADALERFGENRFVSPLSWDLVGRNLFAMAVEGVVFFLITVLIQYRF 1876
Cdd:TIGR01257 1821 tLLRFNAMLRKLLIVFPHFCLGRGLIDLALSQAVTDVYAQFGEEHSANPFQWDLIGKNLVAMAVEGVVYFLLTLLIQHHF 1900
|
1930 1940 1950 1960 1970 1980 1990 2000
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1877 FIRPRPVKAKLPPLNDEDEDVRRERQRILDGGGQNDILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKST 1956
Cdd:TIGR01257 1901 FLSRWIAEPAKEPIFDEDDDVAEERQRIISGGNKTDILRLNELTKVYSGTSSPAVDRLCVGVRPGECFGLLGVNGAGKTT 1980
|
2010 2020 2030 2040 2050 2060 2070 2080
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1957 TFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGL 2036
Cdd:TIGR01257 1981 TFKMLTGDTTVTSGDATVAGKSILTNISDVHQNMGYCPQFDAIDDLLTGREHLYLYARLRGVPAEEIEKVANWSIQSLGL 2060
|
2090 2100 2110 2120 2130 2140 2150 2160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2037 VKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRM 2116
Cdd:TIGR01257 2061 SLYADRLAGTYSGGNKRKLSTAIALIGCPPLVLLDEPTTGMDPQARRMLWNTIVSIIREGRAVVLTSHSMEECEALCTRL 2140
|
2170 2180 2190 2200 2210 2220 2230 2240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2117 AIMVNGRFRCLGSVQHLKNRFGDGYTIVVRIAGSN----PDLKPVQEFFGLAFPGSVLKEKHRNMLQYQLPSslSSLARI 2192
Cdd:TIGR01257 2141 AIMVKGAFQCLGTIQHLKSKFGDGYIVTMKIKSPKddllPDLNPVEQFFQGNFPGSVQRERHYNMLQFQVSS--SSLARI 2218
|
2250 2260 2270 2280
....*....|....*....|....*....|....*....|....*.
gi 568925948 2193 FSILSQSKKRLHIEDYSVSQTTLDQVFVNFAKDQSDDdhlKDLSLH 2238
Cdd:TIGR01257 2219 FQLLISHKDSLLIEEYSVTQTTLDQVFVNFAKQQTET---YDLPLH 2261
|
|
| ABC_subfamily_A |
cd03263 |
ATP-binding cassette domain of the lipid transporters, subfamily A; The ABCA subfamily ... |
1914-2134 |
1.56e-116 |
|
ATP-binding cassette domain of the lipid transporters, subfamily A; The ABCA subfamily mediates the transport of a variety of lipid compounds. Mutations of members of ABCA subfamily are associated with human genetic diseases, such as, familial high-density lipoprotein (HDL) deficiency, neonatal surfactant deficiency, degenerative retinopathies, and congenital keratinization disorders. The ABCA1 protein is involved in disorders of cholesterol transport and high-density lipoprotein (HDL) biosynthesis. The ABCA4 (ABCR) protein transports vitamin A derivatives in the outer segments of photoreceptor cells, and therefore, performs a crucial step in the visual cycle. The ABCA genes are not present in yeast. However, evolutionary studies of ABCA genes indicate that they arose as transporters that subsequently duplicated and that certain sets of ABCA genes were lost in different eukaryotic lineages.
Pssm-ID: 213230 [Multi-domain] Cd Length: 220 Bit Score: 367.99 E-value: 1.56e-116
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03263 1 LQIRNLTKTYKKGTKPAVDDLSLNVYKGEIFGLLGHNGAGKTTTLKMLTGELRPTSGTAYINGYSIRTDRKAARQSLGYC 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03263 81 PQFDALFDELTVREHLRFYARLKGLPKSEIKEEVELLLRVLGLTDKANKRARTLSGGMKRKLSLAIALIGGPSVLLLDEP 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSiVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLK 2134
Cdd:cd03263 161 TSGLDPASRRAIWDLILE-VRKGRSIILTTHSMDEAEALCDRIAIMSDGKLRCIGSPQELK 220
|
|
| ABC_subfamily_A |
cd03263 |
ATP-binding cassette domain of the lipid transporters, subfamily A; The ABCA subfamily ... |
899-1119 |
2.00e-116 |
|
ATP-binding cassette domain of the lipid transporters, subfamily A; The ABCA subfamily mediates the transport of a variety of lipid compounds. Mutations of members of ABCA subfamily are associated with human genetic diseases, such as, familial high-density lipoprotein (HDL) deficiency, neonatal surfactant deficiency, degenerative retinopathies, and congenital keratinization disorders. The ABCA1 protein is involved in disorders of cholesterol transport and high-density lipoprotein (HDL) biosynthesis. The ABCA4 (ABCR) protein transports vitamin A derivatives in the outer segments of photoreceptor cells, and therefore, performs a crucial step in the visual cycle. The ABCA genes are not present in yeast. However, evolutionary studies of ABCA genes indicate that they arose as transporters that subsequently duplicated and that certain sets of ABCA genes were lost in different eukaryotic lineages.
Pssm-ID: 213230 [Multi-domain] Cd Length: 220 Bit Score: 367.60 E-value: 2.00e-116
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03263 1 LQIRNLTKTYKKGTKPAVDDLSLNVYKGEIFGLLGHNGAGKTTTLKMLTGELRPTSGTAYINGYSIRTDRKAARQSLGYC 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03263 81 PQFDALFDELTVREHLRFYARLKGLPKSEIKEEVELLLRVLGLTD-KANKRARTLSGGMKRKLSLAIALIGGPSVLLLDE 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLK 1119
Cdd:cd03263 160 PTSGLDPASRRAIWDLILEVRKGRSIILTTHSMDEAEALCDRIAIMSDGKLRCIGSPQELK 220
|
|
| CcmA |
COG1131 |
ABC-type multidrug transport system, ATPase component [Defense mechanisms]; |
899-1136 |
6.05e-85 |
|
ABC-type multidrug transport system, ATPase component [Defense mechanisms];
Pssm-ID: 440746 [Multi-domain] Cd Length: 236 Bit Score: 278.10 E-value: 6.05e-85
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:COG1131 1 IEVRGLTKRYGD--KTALDGVSLTVEPGEIFGLLGPNGAGKTTTIRMLLGLLRPTSGEVRVLGEDVARDPAEVRRRIGYV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:COG1131 79 PQEPALYPDLTVRENLRFFARLYGLPRKEARERIDELLELFGLTD-AADRKVGTLSGGMKQRLGLALALLHDPELLILDE 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQLGTGYYLTLVKKDVE 1136
Cdd:COG1131 158 PTSGLDPEARRELWELLRELAaEGKTVLLSTHYLEEAERLCDRVAIIDKGRIVADGTPDELKARLLEDVFLELTGEEAR 236
|
|
| CcmA |
COG1131 |
ABC-type multidrug transport system, ATPase component [Defense mechanisms]; |
1914-2137 |
2.82e-77 |
|
ABC-type multidrug transport system, ATPase component [Defense mechanisms];
Pssm-ID: 440746 [Multi-domain] Cd Length: 236 Bit Score: 256.14 E-value: 2.82e-77
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:COG1131 1 IEVRGLTKRY--GDKTALDGVSLTVEPGEIFGLLGPNGAGKTTTIRMLLGLLRPTSGEVRVLGEDVARDPAEVRRRIGYV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:COG1131 79 PQEPALYPDLTVRENLRFFARLYGLPRKEARERIDELLELFGLTDAADRKVGTLSGGMKQRLGLALALLHDPELLILDEP 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRF 2137
Cdd:COG1131 159 TSGLDPEARRELWELLRELAAEGKTVLLSTHYLEEAERLCDRVAIIDKGRIVADGTPDELKARL 222
|
|
| ABC_DR_subfamily_A |
cd03230 |
ATP-binding cassette domain of the drug resistance transporter and related proteins, subfamily ... |
899-1109 |
4.33e-66 |
|
ATP-binding cassette domain of the drug resistance transporter and related proteins, subfamily A; This family of ATP-binding proteins belongs to a multi-subunit transporter involved in drug resistance (BcrA and DrrA), nodulation, lipid transport, and lantibiotic immunity. In bacteria and archaea, these transporters usually include an ATP-binding protein and one or two integral membrane proteins. Eukaryotic systems of the ABCA subfamily display ABC domains that are quite similar to this family. The ATP-binding domain shows the highest similarity between all members of the ABC transporter family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213197 [Multi-domain] Cd Length: 173 Bit Score: 221.50 E-value: 4.33e-66
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03230 1 IEVRNLSKRYGK--KTALDDISLTVEKGEIYGLLGPNGAGKTTLIKIILGLLKPDSGEIKVLGKDIKKEPEEVKRRIGYL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIwfyarlkglsekhvkaemeqmaldvglppsklksktsQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03230 79 PEEPSLYENLTVRENL-------------------------------------KLSGGMKQRLALAQALLHDPELLILDE 121
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03230 122 PTSGLDPESRREFWELLRELKkEGKTILLSSHILEEAERLCDRVAILNNGRI 173
|
|
| NatA |
COG4555 |
ABC-type Na+ transport system, ATPase component NatA [Energy production and conversion, ... |
901-1109 |
2.10e-62 |
|
ABC-type Na+ transport system, ATPase component NatA [Energy production and conversion, Inorganic ion transport and metabolism];
Pssm-ID: 443618 [Multi-domain] Cd Length: 243 Bit Score: 213.95 E-value: 2.10e-62
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQ 980
Cdd:COG4555 4 VENLSKKYGK--VPALKDVSFTAKDGEITGLLGPNGAGKTTLLRMLAGLLKPDSGSILIDGEDVRKEPREARRQIGVLPD 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPT 1060
Cdd:COG4555 82 ERGLYDRLTVRENIRYFAELYGLFDEELKKRIEELIELLGLEE-FLDRRVGELSTGMKKKVALARALVHDPKVLLLDEPT 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 1061 AGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG4555 161 NGLDVMARRLLREILRALKkEGKTVLFSSHIMQEVEALCDRVVILHKGKV 210
|
|
| ABC_DrrA |
cd03265 |
Daunorubicin/doxorubicin resistance ATP-binding protein; DrrA is the ATP-binding protein ... |
899-1119 |
6.07e-61 |
|
Daunorubicin/doxorubicin resistance ATP-binding protein; DrrA is the ATP-binding protein component of a bacterial exporter complex that confers resistance to the antibiotics daunorubicin and doxorubicin. In addition to DrrA, the complex includes an integral membrane protein called DrrB. DrrA belongs to the ABC family of transporters and shares sequence and functional similarities with a protein found in cancer cells called P-glycoprotein. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region in addition to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213232 [Multi-domain] Cd Length: 220 Bit Score: 208.76 E-value: 6.07e-61
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03265 1 IEVENLVKKY--GDFEAVRGVSFRVRRGEIFGLLGPNGAGKTTTIKMLTTLLKPTSGRATVAGHDVVREPREVRRRIGIV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKsKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03265 79 FQDLSVDDELTGWENLYIHARLYGVPGAERRERIDELLDFVGLLEAADR-LVKTYSGGMRRRLEIARSLVHRPEVLFLDE 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1059 PTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLK 1119
Cdd:cd03265 158 PTIGLDPQTRAHVWEYIekLKEEFGMTILLTTHYMEEAEQLCDRVAIIDHGRIIAEGTPEELK 220
|
|
| ABC_DrrA |
cd03265 |
Daunorubicin/doxorubicin resistance ATP-binding protein; DrrA is the ATP-binding protein ... |
1914-2134 |
2.95e-59 |
|
Daunorubicin/doxorubicin resistance ATP-binding protein; DrrA is the ATP-binding protein component of a bacterial exporter complex that confers resistance to the antibiotics daunorubicin and doxorubicin. In addition to DrrA, the complex includes an integral membrane protein called DrrB. DrrA belongs to the ABC family of transporters and shares sequence and functional similarities with a protein found in cancer cells called P-glycoprotein. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region in addition to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213232 [Multi-domain] Cd Length: 220 Bit Score: 203.76 E-value: 2.95e-59
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03265 1 IEVENLVKKYGDFE--AVRGVSFRVRRGEIFGLLGPNGAGKTTTIKMLTTLLKPTSGRATVAGHDVVREPREVRRRIGIV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03265 79 FQDLSVDDELTGWENLYIHARLYGVPGAERRERIDELLDFVGLLEAADRLVKTYSGGMRRRLEIARSLVHRPEVLFLDEP 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLK 2134
Cdd:cd03265 159 TIGLDPQTRAHVWEYIEKLKEEfGMTILLTTHYMEEAEQLCDRVAIIDHGRIIAEGTPEELK 220
|
|
| drrA |
TIGR01188 |
daunorubicin resistance ABC transporter ATP-binding subunit; This model describes daunorubicin ... |
906-1123 |
5.03e-59 |
|
daunorubicin resistance ABC transporter ATP-binding subunit; This model describes daunorubicin resistance ABC transporter, ATP binding subunit in bacteria and archaea. This model is restricted in its scope to preferentially recognize the ATP binding subunit associated with effux of the drug, daunorubicin. This transport system belong to the larger ATP-Binding Cassette (ABC) transporter superfamily. The characteristic feature of these transporter is the obligatory coupling of ATP hydrolysis to substrate translocation. The minimal configuration of bacterial ABC transport system: an ATPase or ATP binding subunit; An integral membrane protein; a hydrophilic polypetpide, which likely functions as substrate binding protein. In eukaryotes proteins of similar function include p-gyco proteins, multidrug resistance protein etc. [Transport and binding proteins, Other]
Pssm-ID: 130256 [Multi-domain] Cd Length: 302 Bit Score: 206.47 E-value: 5.03e-59
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 906 KVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLF 985
Cdd:TIGR01188 1 KVY--GDFKAVDGVNFKVREGEVFGFLGPNGAGKTTTIRMLTTLLRPTSGTARVAGYDVVREPRKVRRSIGIVPQYASVD 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 DMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:TIGR01188 79 EDLTGRENLEMMGRLYGLPKDEAEERAEELLELFELG-EAADRPVGTYSGGMRRRLDIAASLIHQPDVLFLDEPTTGLDP 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1066 YSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQLG 1123
Cdd:TIGR01188 158 RTRRAIWDYIRALKEeGVTILLTTHYMEEADKLCDRIAIIDHGRIIAEGTPEELKRRLG 216
|
|
| drrA |
TIGR01188 |
daunorubicin resistance ABC transporter ATP-binding subunit; This model describes daunorubicin ... |
1929-2222 |
2.01e-58 |
|
daunorubicin resistance ABC transporter ATP-binding subunit; This model describes daunorubicin resistance ABC transporter, ATP binding subunit in bacteria and archaea. This model is restricted in its scope to preferentially recognize the ATP binding subunit associated with effux of the drug, daunorubicin. This transport system belong to the larger ATP-Binding Cassette (ABC) transporter superfamily. The characteristic feature of these transporter is the obligatory coupling of ATP hydrolysis to substrate translocation. The minimal configuration of bacterial ABC transport system: an ATPase or ATP binding subunit; An integral membrane protein; a hydrophilic polypetpide, which likely functions as substrate binding protein. In eukaryotes proteins of similar function include p-gyco proteins, multidrug resistance protein etc. [Transport and binding proteins, Other]
Pssm-ID: 130256 [Multi-domain] Cd Length: 302 Bit Score: 204.55 E-value: 2.01e-58
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQFDAITELLTGREH 2008
Cdd:TIGR01188 7 KAVDGVNFKVREGEVFGFLGPNGAGKTTTIRMLTTLLRPTSGTARVAGYDVVREPRKVRRSIGIVPQYASVDEDLTGREN 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2009 VEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNC 2088
Cdd:TIGR01188 87 LEMMGRLYGLPKDEAEERAEELLELFELGEAADRPVGTYSGGMRRRLDIAASLIHQPDVLFLDEPTTGLDPRTRRAIWDY 166
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2089 ALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIVVRIAGSnpDLKPVQEFFGLAFPGS 2168
Cdd:TIGR01188 167 IRALKEEGVTILLTTHYMEEADKLCDRIAIIDHGRIIAEGTPEELKRRLGKDTLESRPRDIQ--SLKVEVSMLIAELGET 244
|
250 260 270 280 290
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2169 VL--KEKHRNMLQYQLPSSLSS--LARIFSILsqSKKRLHIEDYSVSQTTLDQVFVNF 2222
Cdd:TIGR01188 245 GLglLAVTVDSDRIKILVPDGDetVPEIVEAA--IRNGIRIRSISTERPSLDDVFLKL 300
|
|
| ABC_DR_subfamily_A |
cd03230 |
ATP-binding cassette domain of the drug resistance transporter and related proteins, subfamily ... |
1914-2124 |
3.15e-57 |
|
ATP-binding cassette domain of the drug resistance transporter and related proteins, subfamily A; This family of ATP-binding proteins belongs to a multi-subunit transporter involved in drug resistance (BcrA and DrrA), nodulation, lipid transport, and lantibiotic immunity. In bacteria and archaea, these transporters usually include an ATP-binding protein and one or two integral membrane proteins. Eukaryotic systems of the ABCA subfamily display ABC domains that are quite similar to this family. The ATP-binding domain shows the highest similarity between all members of the ABC transporter family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213197 [Multi-domain] Cd Length: 173 Bit Score: 196.08 E-value: 3.15e-57
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03230 1 IEVRNLSKRY--GKKTALDDISLTVEKGEIYGLLGPNGAGKTTLIKIILGLLKPDSGEIKVLGKDIKKEPEEVKRRIGYL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEffallrgvpekevgkvgewairklglvkygekyasnYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03230 79 PEEPSLYENLTVRENLK------------------------------------LSGGMKQRLALAQALLHDPELLILDEP 122
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:cd03230 123 TSGLDPESRREFWELLRELKKEGKTILLSSHILEEAERLCDRVAILNNGRI 173
|
|
| ABC_drug_resistance_like |
cd03264 |
ABC-type multidrug transport system, ATPase component; The biological function of this family ... |
899-1109 |
2.64e-55 |
|
ABC-type multidrug transport system, ATPase component; The biological function of this family is not well characterized, but display ABC domains similar to members of ABCA subfamily. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213231 [Multi-domain] Cd Length: 211 Bit Score: 192.02 E-value: 2.64e-55
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMkvAVDGLALNFYEGqITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03264 1 LQLENLTKRYGKKR--ALDGVSLTLGPG-MYGLLGPNGAGKTTLMRILATLTPPSSGTIRIDGQDVLKQPQKLRRRIGYL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03264 78 PQEFGVYPNFTVREFLDYIAWLKGIPSKEVKARVDEVLELVNLGDRA-KKKIGSLSGGMRRRVGIAQALVGDPSILIVDE 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03264 157 PTAGLDPEERIRFRNLLSELGEDRIVILSTHIVEDVESLCNQVAVLNKGKL 207
|
|
| NatA |
COG4555 |
ABC-type Na+ transport system, ATPase component NatA [Energy production and conversion, ... |
1914-2139 |
8.86e-54 |
|
ABC-type Na+ transport system, ATPase component NatA [Energy production and conversion, Inorganic ion transport and metabolism];
Pssm-ID: 443618 [Multi-domain] Cd Length: 243 Bit Score: 188.91 E-value: 8.86e-54
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:COG4555 2 IEVENLSKKY--GKVPALKDVSFTAKDGEITGLLGPNGAGKTTLLRMLAGLLKPDSGSILIDGEDVRKEPREARRQIGVL 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:COG4555 80 PDERGLYDRLTVRENIRYFAELYGLFDEELKKRIEELIELLGLEEFLDRRVGELSTGMKKKVALARALVHDPKVLLLDEP 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGD 2139
Cdd:COG4555 160 TNGLDVMARRLLREILRALKKEGKTVLFSSHIMQEVEALCDRVVILHKGKVVAQGSLDELREEIGE 225
|
|
| YhaQ |
COG4152 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
899-1108 |
2.40e-48 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 443322 [Multi-domain] Cd Length: 298 Bit Score: 175.30 E-value: 2.40e-48
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEmssIRQNLGVC 978
Cdd:COG4152 2 LELKGLTKRFGD--KTAVDDVSFTVPKGEIFGLLGPNGAGKTTTIRIILGILAPDSGEVLWDGEPLDPE---DRRRIGYL 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:COG4152 77 PEERGLYPKMKVGEQLVYLARLKGLSKAEAKRRADEWLERLGL-GDRANKKVEELSKGNQQKVQLIAALLHDPELLILDE 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:COG4152 156 PFSGLDPVNVELLKDVIRELAaKGTTVIFSSHQMELVEELCDRIVIINKGR 206
|
|
| ABC_cobalt_CbiO_domain1 |
cd03225 |
First domain of the ATP-binding cassette component of cobalt transport system; Domain I of the ... |
900-1108 |
3.52e-48 |
|
First domain of the ATP-binding cassette component of cobalt transport system; Domain I of the ABC component of a cobalt transport family found in bacteria, archaea, and eukaryota. The transition metal cobalt is an essential component of many enzymes and must be transported into cells in appropriate amounts when needed. This ABC transport system of the CbiMNQO family is involved in cobalt transport in association with the cobalamin (vitamin B12) biosynthetic pathways. Most of cobalt (Cbi) transport systems possess a separate CbiN component, the cobalt-binding periplasmic protein, and they are encoded by the conserved gene cluster cbiMNQO. Both the CbiM and CbiQ proteins are integral cytoplasmic membrane proteins, and the CbiO protein has the linker peptide and the Walker A and B motifs commonly found in the ATPase components of the ABC-type transport systems.
Pssm-ID: 213192 [Multi-domain] Cd Length: 211 Bit Score: 171.50 E-value: 3.52e-48
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGVC 978
Cdd:cd03225 1 ELKNLSFSYPDGARPALDDISLTIKKGEFVLIVGPNGSGKSTLLRLLNGLLGPTSGEVLVDGKDLTKLsLKELRRKVGLV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:cd03225 81 FQNpDDQFFGPTVEEEVAFGLENLGLPEEEIEERVEEALELVGL-EGLRDRSPFTLSGGQKQRVAIAGVLAMDPDILLLD 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd03225 160 EPTAGLDPAGRRELLELLKKLkAEGKTIIIVTHDLDLLLELADRVIVLEDGK 211
|
|
| EcfA2 |
COG1122 |
Energy-coupling factor transporter ATP-binding protein EcfA2 [Inorganic ion transport and ... |
899-1114 |
2.07e-47 |
|
Energy-coupling factor transporter ATP-binding protein EcfA2 [Inorganic ion transport and metabolism, General function prediction only];
Pssm-ID: 440739 [Multi-domain] Cd Length: 230 Bit Score: 170.21 E-value: 2.07e-47
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGV 977
Cdd:COG1122 1 IELENLSFSYPGG-TPALDDVSLSIEKGEFVAIIGPNGSGKSTLLRLLNGLLKPTSGEVLVDGKDITKKnLRELRRKVGL 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQH--NVLFdMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLppSKLKSK-TSQLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:COG1122 80 VFQNpdDQLF-APTVEEDVAFGPENLGLPREEIRERVEEALELVGL--EHLADRpPHELSGGQKQRVAIAGVLAMEPEVL 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:COG1122 157 VLDEPTAGLDPRGRRELLELLKRLnKEGKTVIIVTHDLDLVAELADRVIVLDDGRIVADGT 217
|
|
| ABC_NatA_sodium_exporter |
cd03266 |
ATP-binding cassette domain of the Na+ transporter; NatA is the ATPase component of a ... |
899-1113 |
4.77e-47 |
|
ATP-binding cassette domain of the Na+ transporter; NatA is the ATPase component of a bacterial ABC-type Na+ transport system called NatAB, which catalyzes ATP-dependent electrogenic Na+ extrusion without mechanically coupled proton or K+ uptake. NatB possess six putative membrane spanning regions at its C-terminus. In B. subtilis, NatAB is inducible by agents such as ethanol and protonophores, which lower the proton-motive force across the membrane. The closest sequence similarity to NatA is exhibited by DrrA of the two-component daunorubicin- and doxorubicin-efflux system. Hence, the functional NatAB is presumably assembled with two copies of a single ATP-binding protein and a single integral membrane protein.
Pssm-ID: 213233 [Multi-domain] Cd Length: 218 Bit Score: 168.70 E-value: 4.77e-47
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLG 976
Cdd:cd03266 2 ITADALTKRFRDVKKtvQAVDGVSFTVKPGEVTGLLGPNGAGKTTTLRMLAGLLEPDAGFATVDGFDVVKEPAEARRRLG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSkLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03266 82 FVSDSTGLYDRLTARENLEYFAGLYGLKGDELTARLEELADRLGMEEL-LDRRVGGFSTGMRQKVAIARALVHDPPVLLL 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03266 161 DEPTTGLDVMATRALREFIRQLRaLGKCILFSTHIMQEVERLCDRVVVLHRGRVVYEG 218
|
|
| ABC_drug_resistance_like |
cd03264 |
ABC-type multidrug transport system, ATPase component; The biological function of this family ... |
1914-2125 |
9.65e-47 |
|
ABC-type multidrug transport system, ATPase component; The biological function of this family is not well characterized, but display ABC domains similar to members of ABCA subfamily. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213231 [Multi-domain] Cd Length: 211 Bit Score: 167.76 E-value: 9.65e-47
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGeCFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03264 1 LQLENLTKRYGKKR--ALDGVSLTLGPG-MYGLLGPNGAGKTTLMRILATLTPPSSGTIRIDGQDVLKQPQKLRRRIGYL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03264 78 PQEFGVYPNFTVREFLDYIAWLKGIPSKEVKARVDEVLELVNLGDRAKKKIGSLSGGMRRRVGIAQALVGDPSILIVDEP 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2074 TTGMDPKAR-RF--LwncaLSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFR 2125
Cdd:cd03264 158 TAGLDPEERiRFrnL----LSELGEDRIVILSTHIVEDVESLCNQVAVLNKGKLV 208
|
|
| ABC_BcrA_bacitracin_resist |
cd03268 |
ATP-binding cassette domain of the bacitracin-resistance transporter; The BcrA subfamily ... |
899-1109 |
3.06e-46 |
|
ATP-binding cassette domain of the bacitracin-resistance transporter; The BcrA subfamily represents ABC transporters involved in peptide antibiotic resistance. Bacitracin is a dodecapeptide antibiotic produced by B. licheniformis and B. subtilis. The synthesis of bacitracin is non-ribosomally catalyzed by a multi-enzyme complex BcrABC. Bacitracin has potent antibiotic activity against gram-positive bacteria. The inhibition of peptidoglycan biosynthesis is the best characterized bacterial effect of bacitracin. The bacitracin resistance of B. licheniformis is mediated by the ABC transporter Bcr which is composed of two identical BcrA ATP-binding subunits and one each of the integral membrane proteins, BcrB and BcrC. B. subtilis cells carrying bcr genes on high-copy number plasmids develop collateral detergent sensitivity, a similar phenomenon in human cells with overexpressed multi-drug resistance P-glycoprotein.
Pssm-ID: 213235 [Multi-domain] Cd Length: 208 Bit Score: 165.85 E-value: 3.06e-46
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:cd03268 1 LKTNDLTKTY--GKKRVLDDISLHVKKGEIYGFLGPNGAGKTTTMKIILGLIKPDSGEITFDGKSY-QKNIEALRRIGAL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVkaemEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03268 78 IEAPGFYPNLTARENLRLLARLLGIRKKRI----DEVLDVVGLKDSA-KKKVKGFSLGMKQRLGIALALLGNPDLLILDE 152
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03268 153 PTNGLDPDGIKELRELILSLRdQGITVLISSHLLSEIQKVADRIGIINKGKL 204
|
|
| ABC_tran |
pfam00005 |
ABC transporter; ABC transporters for a large family of proteins responsible for translocation ... |
916-1061 |
1.49e-44 |
|
ABC transporter; ABC transporters for a large family of proteins responsible for translocation of a variety of compounds across biological membranes. ABC transporters are the largest family of proteins in many completely sequenced bacteria. ABC transporters are composed of two copies of this domain and two copies of a transmembrane domain pfam00664. These four domains may belong to a single polypeptide or belong in different polypeptide chains.
Pssm-ID: 394964 [Multi-domain] Cd Length: 150 Bit Score: 158.97 E-value: 1.49e-44
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMLTVEEHI 994
Cdd:pfam00005 1 LKNVSLTLNPGEILALVGPNGAGKSTLLKLIAGLLSPTEGTILLDGQDLTDdERKSLRKEIGYVFQDPQLFPRLTVRENL 80
|
90 100 110 120 130 140 150
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 995 WFYARLKGLSEKHVKAEMEQMALDVGLPP---SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTA 1061
Cdd:pfam00005 81 RLGLLLKGLSKREKDARAEEALEKLGLGDladRPVGERPGTLSGGQRQRVAIARALLTKPKLLLLDEPTA 150
|
|
| ABC_Carb_Solutes_like |
cd03259 |
ATP-binding cassette domain of the carbohydrate and solute transporters-like; This family is ... |
899-1109 |
2.34e-44 |
|
ATP-binding cassette domain of the carbohydrate and solute transporters-like; This family is comprised of proteins involved in the transport of apparently unrelated solutes and proteins specific for di- and oligosaccharides and polyols. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213226 [Multi-domain] Cd Length: 213 Bit Score: 160.76 E-value: 2.34e-44
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:cd03259 1 LELKGLSKTYGS--VRALDDLSLTVEPGEFLALLGPSGCGKTTLLRLIAGLERPDSGEILIDGRDV-TGVPPERRNIGMV 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03259 78 FQDYALFPHLTVAENIAFGLKLRGVPKAEIRARVRELLELVGLEG-LLNRYPHELSGGQQQRVALARALAREPSLLLLDE 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1059 PTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03259 157 PLSALDAKLREELREELKELqrELGITTIYVTHDQEEALALADRIAVMNEGRI 209
|
|
| ABC_OpuCA_Osmoprotection |
cd03295 |
ATP-binding cassette domain of the osmoprotectant transporter; OpuCA is a the ATP binding ... |
899-1114 |
2.72e-44 |
|
ATP-binding cassette domain of the osmoprotectant transporter; OpuCA is a the ATP binding component of a bacterial solute transporter that serves a protective role to cells growing in a hyperosmolar environment. ABC (ATP-binding cassette) transporter nucleotide-binding domain; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition, to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213262 [Multi-domain] Cd Length: 242 Bit Score: 161.70 E-value: 2.72e-44
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGV 977
Cdd:cd03295 1 IEFENVTKRYGGG-KKAVNNLNLEIAKGEFLVLIGPSGSGKTTTMKMINRLIEPTSGEIFIDGEDIREQdPVELRRKIGY 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSK-TSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03295 80 VIQQIGLFPHMTVEENIALVPKLLKWPKEKIRERADELLALVGLDPAEFADRyPHELSGGQQQRVGVARALAADPPLLLM 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:cd03295 160 DEPFGALDPITRDQLQEEFKRLQQelGKTIVFVTHDIDEAFRLADRIAIMKNGEIVQVGT 219
|
|
| ABC_Org_Solvent_Resistant |
cd03261 |
ATP-binding cassette transport system involved in resistance to organic solvents; ABC ... |
899-1114 |
2.90e-44 |
|
ATP-binding cassette transport system involved in resistance to organic solvents; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213228 [Multi-domain] Cd Length: 235 Bit Score: 161.52 E-value: 2.90e-44
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQN 974
Cdd:cd03261 1 IELRGLTKSFGG--RTVLKGVDLDVRRGEILAIIGPSGSGKSTLLRLIVGLLRPDSGEVLIDGEDIsglsEAELYRLRRR 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALD-VGLPPSKLKsKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:cd03261 79 MGMLFQSGALFDSLTVFENVAFPLREHTRLSEEEIREIVLEKLEaVGLRGAEDL-YPAELSGGMKKRVALARALALDPEL 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:cd03261 158 LLYDEPTAGLDPIASGVIDDLIRSLKKelGLTSIMVTHDLDTAFAIADRIAVLYDGKIVAEGT 220
|
|
| MlaF |
COG1127 |
ATPase subunit MlaF of the ABC-type intermembrane phospholipid transporter Mla [Cell wall ... |
899-1114 |
4.69e-44 |
|
ATPase subunit MlaF of the ABC-type intermembrane phospholipid transporter Mla [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440744 [Multi-domain] Cd Length: 241 Bit Score: 160.92 E-value: 4.69e-44
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQN 974
Cdd:COG1127 6 IEVRNLTKSFGD--RVVLDGVSLDVPRGEILAIIGGSGSGKSVLLKLIIGLLRPDSGEILVDGQDItglsEKELYELRRR 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHNVLFDMLTVEEHIWFYAR-LKGLSEkhvkAEMEQMALD----VGLPPSKLKsKTSQLSGGMQRKLSVALAFVG 1049
Cdd:COG1127 84 IGMLFQGGALFDSLTVFENVAFPLReHTDLSE----AEIRELVLEklelVGLPGAADK-MPSELSGGMRKRVALARALAL 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1050 GSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:COG1127 159 DPEILLYDEPTAGLDPITSAVIDELIRELRDelGLTSVVVTHDLDSAFAIADRVAVLADGKIIAEGT 225
|
|
| ABC_NatA_sodium_exporter |
cd03266 |
ATP-binding cassette domain of the Na+ transporter; NatA is the ATPase component of a ... |
1913-2128 |
4.70e-43 |
|
ATP-binding cassette domain of the Na+ transporter; NatA is the ATPase component of a bacterial ABC-type Na+ transport system called NatAB, which catalyzes ATP-dependent electrogenic Na+ extrusion without mechanically coupled proton or K+ uptake. NatB possess six putative membrane spanning regions at its C-terminus. In B. subtilis, NatAB is inducible by agents such as ethanol and protonophores, which lower the proton-motive force across the membrane. The closest sequence similarity to NatA is exhibited by DrrA of the two-component daunorubicin- and doxorubicin-efflux system. Hence, the functional NatAB is presumably assembled with two copies of a single ATP-binding protein and a single integral membrane protein.
Pssm-ID: 213233 [Multi-domain] Cd Length: 218 Bit Score: 157.14 E-value: 4.70e-43
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNM 1990
Cdd:cd03266 1 MITADALTKRFRDVKKTvqAVDGVSFTVKPGEVTGLLGPNGAGKTTTLRMLAGLLEPDAGFATVDGFDVVKEPAEARRRL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:cd03266 81 GFVSDSTGLYDRLTARENLEYFAGLYGLKGDELTARLEELADRLGMEELLDRRVGGFSTGMRQKVAIARALVHDPPVLLL 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:cd03266 161 DEPTTGLDVMATRALREFIRQLRALGKCILFSTHIMQEVERLCDRVVVLHRGRVVYEG 218
|
|
| ABC_NrtD_SsuB_transporters |
cd03293 |
ATP-binding cassette domain of the nitrate and sulfonate transporters; NrtD and SsuB are the ... |
899-1105 |
1.08e-42 |
|
ATP-binding cassette domain of the nitrate and sulfonate transporters; NrtD and SsuB are the ATP-binding subunits of the bacterial ABC-type nitrate and sulfonate transport systems, respectively. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213260 [Multi-domain] Cd Length: 220 Bit Score: 156.09 E-value: 1.08e-42
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDG--MKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRsemsSIRQNLG 976
Cdd:cd03293 1 LEVRNVSKTYGGGggAVTALEDISLSVEEGEFVALVGPSGCGKSTLLRIIAGLERPTSGEVLVDGEPVT----GPGPDRG 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSkLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03293 77 YVFQQDALLPWLTVLDNVALGLELQGVPKAEARERAEELLELVGLSGF-ENAYPHQLSGGMRQRVALARALAVDPDVLLL 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLK--YRQGRTIILSTHHMDEADILGDRIAIIS 1105
Cdd:cd03293 156 DEPFSALDALTREQLQEELLDiwRETGKTVLLVTHDIDEAVFLADRVVVLS 206
|
|
| ABC_BcrA_bacitracin_resist |
cd03268 |
ATP-binding cassette domain of the bacitracin-resistance transporter; The BcrA subfamily ... |
1914-2123 |
1.84e-42 |
|
ATP-binding cassette domain of the bacitracin-resistance transporter; The BcrA subfamily represents ABC transporters involved in peptide antibiotic resistance. Bacitracin is a dodecapeptide antibiotic produced by B. licheniformis and B. subtilis. The synthesis of bacitracin is non-ribosomally catalyzed by a multi-enzyme complex BcrABC. Bacitracin has potent antibiotic activity against gram-positive bacteria. The inhibition of peptidoglycan biosynthesis is the best characterized bacterial effect of bacitracin. The bacitracin resistance of B. licheniformis is mediated by the ABC transporter Bcr which is composed of two identical BcrA ATP-binding subunits and one each of the integral membrane proteins, BcrB and BcrC. B. subtilis cells carrying bcr genes on high-copy number plasmids develop collateral detergent sensitivity, a similar phenomenon in human cells with overexpressed multi-drug resistance P-glycoprotein.
Pssm-ID: 213235 [Multi-domain] Cd Length: 208 Bit Score: 155.07 E-value: 1.84e-42
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNiHEVHQNMGYC 1993
Cdd:cd03268 1 LKTNDLTKTY--GKKRVLDDISLHVKKGEIYGFLGPNGAGKTTTMKIILGLIKPDSGEITFDGKSYQKN-IEALRRIGAL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVgewaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03268 78 IEAPGFYPNLTARENLRLLARLLGIRKKRIDEV----LDVVGLKDSAKKKVKGFSLGMKQRLGIALALLGNPDLLILDEP 153
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03268 154 TNGLDPDGIKELRELILSLRDQGITVLISSHLLSEIQKVADRIGIINKGK 203
|
|
| ABC_putative_ATPase |
cd03269 |
ATP-binding cassette domain of an uncharacterized transporter; This subgroup is related to the ... |
899-1108 |
2.47e-42 |
|
ATP-binding cassette domain of an uncharacterized transporter; This subgroup is related to the subfamily A transporters involved in drug resistance, nodulation, lipid transport, and bacteriocin and lantibiotic immunity. In eubacteria and archaea, the typical organization consists of one ABC and one or two integral membranes. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region in addition to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213236 [Multi-domain] Cd Length: 210 Bit Score: 154.75 E-value: 2.47e-42
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEmssIRQNLGVC 978
Cdd:cd03269 1 LEVENVTKRFGR--VTALDDISFSVEKGEIFGLLGPNGAGKTTTIRMILGIILPDSGEVLFDGKPLDIA---ARNRIGYL 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03269 76 PEERGLYPKMKVIDQLVYLAQLKGLKKEEARRRIDEWLERLELSE-YANKRVEELSKGNQQKVQFIAAVIHDPELLILDE 154
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd03269 155 PFSGLDPVNVELLKDVIRELaRAGKTVILSTHQMELVEELCDRVLLLNKGR 205
|
|
| ABC_MJ0796_LolCDE_FtsE |
cd03255 |
ATP-binding cassette domain of the transporters involved in export of lipoprotein and ... |
899-1109 |
2.80e-42 |
|
ATP-binding cassette domain of the transporters involved in export of lipoprotein and macrolide, and Cell division ATP-binding protein FtsE; This family is comprised of MJ0796 ATP-binding cassette, macrolide-specific ABC-type efflux carrier (MacAB), and proteins involved in cell division (FtsE), and release of lipoproteins from the cytoplasmic membrane (LolCDE). They are clustered together phylogenetically. MacAB is an exporter that confers resistance to macrolides, while the LolCDE system is not a transporter at all. The FtsEX complex resembles an ABC transporter, where FtsE is the ATPase and the membrane subunit FtsX resembles a permease subunit. But rather than transporting any substrate, the complex acts in cell division by undergoing conformational changes that alter the activity of cell wall hydrolases located outside the plasma membrane. The complex is widely conserved in bacteria, but also extremely divergent in sequence between different lineages. The LolCDE complex catalyzes the release of lipoproteins from the cytoplasmic membrane prior to their targeting to the outer membrane.
Pssm-ID: 213222 [Multi-domain] Cd Length: 218 Bit Score: 154.96 E-value: 2.80e-42
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:cd03255 1 IELKNLSKTYGGGGEkvQALKGVSLSIEKGEFVAIVGPSGSGKSTLLNILGGLDRPTSGEVRVDGTDIsklsEKELAAFR 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 -QNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:cd03255 81 rRHIGFVFQSFNLLPDLTALENVELPLLLAGVPKKERRERAEELLERVGL-GDRLNHYPSELSGGQQQRVAIARALANDP 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADiLGDRIAIISHGKL 1109
Cdd:cd03255 160 KIILADEPTGNLDSETGKEVMELLRELNKeaGTTIVVVTHDPELAE-YADRIIELRDGKI 218
|
|
| CcmA |
COG4133 |
ABC-type transport system involved in cytochrome c biogenesis, ATPase component ... |
899-1101 |
4.47e-41 |
|
ABC-type transport system involved in cytochrome c biogenesis, ATPase component [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 443308 [Multi-domain] Cd Length: 206 Bit Score: 151.09 E-value: 4.47e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:COG4133 3 LEAENLSCRRGE--RLLFSGLSFTLAAGEALALTGPNGSGKTTLLRILAGLLPPSAGEVLWNGEPIRDAREDYRRRLAYL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLsekHVKAEMEQMALD-VGLPPSkLKSKTSQLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:COG4133 81 GHADGLKPELTVRENLRFWAALYGL---RADREAIDEALEaVGLAGL-ADLPVRQLSAGQKRRVALARLLLSPAPLWLLD 156
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHhmDEADILGDRI 1101
Cdd:COG4133 157 EPFTALDAAGVALLAELIAAHLaRGGAVLLTTH--QPLELAAARV 199
|
|
| PRK13536 |
PRK13536 |
nodulation factor ABC transporter ATP-binding protein NodI; |
897-1108 |
5.09e-41 |
|
nodulation factor ABC transporter ATP-binding protein NodI;
Pssm-ID: 237419 [Multi-domain] Cd Length: 340 Bit Score: 155.76 E-value: 5.09e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLG 976
Cdd:PRK13536 40 VAIDLAGVSKSYGD--KAVVNGLSFTVASGECFGLLGPNGAGKSTIARMILGMTSPDAGKITVLGVPVPARARLARARIG 117
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMaLDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:PRK13536 118 VVPQFDNLDLEFTVRENLLVFGRYFGMSTREIEAVIPSL-LEFARLESKADARVSDLSGGMKRRLTLARALINDPQLLIL 196
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK13536 197 DEPTTGLDPHARHLIWERLRSlLARGKTILLTTHFMEEAERLCDRLCVLEAGR 249
|
|
| PRK13537 |
PRK13537 |
nodulation factor ABC transporter ATP-binding protein NodI; |
899-1108 |
5.97e-41 |
|
nodulation factor ABC transporter ATP-binding protein NodI;
Pssm-ID: 237420 [Multi-domain] Cd Length: 306 Bit Score: 154.19 E-value: 5.97e-41
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:PRK13537 8 IDFRNVEKRY--GDKLVVDGLSFHVQRGECFGLLGPNGAGKTTTLRMLLGLTHPDAGSISLCGEPVPSRARHARQRVGVV 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMeQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:PRK13537 86 PQFDNLDPDFTVRENLLVFGRYFGLSAAAARALV-PPLLEFAKLENKADAKVGELSGGMKRRLTLARALVNDPDVLVLDE 164
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK13537 165 PTTGLDPQARHLMWERLRSlLARGKTILLTTHFMEEAERLCDRLCVIEEGR 215
|
|
| TauB |
COG1116 |
ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component [Inorganic ion ... |
888-1107 |
3.58e-40 |
|
ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 440733 [Multi-domain] Cd Length: 260 Bit Score: 150.63 E-value: 3.58e-40
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 888 MEEEPTHLRlgvsIQNLVKVY--RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR 965
Cdd:COG1116 1 MSAAAPALE----LRGVSKRFptGGGGVTALDDVSLTVAAGEFVALVGPSGCGKSTLLRLIAGLEKPTSGEVLVDGKPVT 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 966 SemssIRQNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVAL 1045
Cdd:COG1116 77 G----PGPDRGVVFQEPALLPWLTVLDNVALGLELRGVPKAERRERARELLELVGLAG-FEDAYPHQLSGGMRQRVAIAR 151
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1046 AFVGGSKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHG 1107
Cdd:COG1116 152 ALANDPEVLLMDEPFGALDALTRERLQDELLRLwqETGKTVLFVTHDVDEAVFLADRVVVLSAR 215
|
|
| YhaQ |
COG4152 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
1913-2228 |
6.90e-40 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 443322 [Multi-domain] Cd Length: 298 Bit Score: 151.03 E-value: 6.90e-40
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlsnIHEVHQNMGY 1992
Cdd:COG4152 1 MLELKGLTKRFGDKT--AVDDVSFTVPKGEIFGLLGPNGAGKTTTIRIILGILAPDSGEVLWDGEPL---DPEDRRRIGY 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPqfdaitE---L---LTGREHVEFFALLRGVPEKEVGK-VGEWaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:COG4152 76 LP------EergLypkMKVGEQLVYLARLKGLSKAEAKRrADEW-LERLGLGDRANKKVEELSKGNQQKVQLIAALLHDP 148
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGdGYTIVV 2145
Cdd:COG4152 149 ELLILDEPFSGLDPVNVELLKDVIRELAAKGTTVIFSSHQMELVEELCDRIVIINKGRKVLSGSVDEIRRQFG-RNTLRL 227
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2146 RIAGSNPDLKpvqeffglAFPGSVLKEKHRNMLQYQLPSSLSSlARIFSILSQskkRLHIEDYSVSQTTLDQVFVNFAKD 2225
Cdd:COG4152 228 EADGDAGWLR--------ALPGVTVVEEDGDGAELKLEDGADA-QELLRALLA---RGPVREFEEVRPSLNEIFIEVVGE 295
|
...
gi 568925948 2226 QSD 2228
Cdd:COG4152 296 KAE 298
|
|
| ABC_ATPase |
cd00267 |
ATP-binding cassette transporter nucleotide-binding domain; ABC transporters are a large ... |
900-1108 |
1.75e-39 |
|
ATP-binding cassette transporter nucleotide-binding domain; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213179 [Multi-domain] Cd Length: 157 Bit Score: 144.69 E-value: 1.75e-39
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS-IRQNLGVC 978
Cdd:cd00267 1 EIENLSFRYGG--RTALDNVSLTLKAGEIVALVGPNGSGKSTLLRAIAGLLKPTSGEILIDGKDIAKLPLEeLRRRIGYV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQhnvlfdmltveehiwfyarlkglsekhvkaemeqmaldvglppsklksktsqLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd00267 79 PQ----------------------------------------------------LSGGQRQRVALARALLLNPDLLLLDE 106
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd00267 107 PTSGLDPASRERLLELLRELAEeGRTVIIVTHDPELAELAADRVIVLKDGK 157
|
|
| LolD |
COG1136 |
ABC-type lipoprotein export system, ATPase component [Cell wall/membrane/envelope biogenesis]; |
899-1109 |
4.04e-39 |
|
ABC-type lipoprotein export system, ATPase component [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440751 [Multi-domain] Cd Length: 227 Bit Score: 146.34 E-value: 4.04e-39
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:COG1136 5 LELRNLTKSYGTGEGevTALRGVSLSIEAGEFVAIVGPSGSGKSTLLNILGGLDRPTSGEVLIDGQDIsslsERELARLR 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 -QNLGVCPQ-HNvLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGG 1050
Cdd:COG1136 85 rRHIGFVFQfFN-LLPELTALENVALPLLLAGVSRKERRERARELLERVGLG-DRLDHRPSQLSGGQQQRVAIARALVNR 162
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1051 SKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADIlGDRIAIISHGKL 1109
Cdd:COG1136 163 PKLILADEPTGNLDSKTGEEVLELLRELnrELGTTIVMVTHDPELAAR-ADRVIRLRDGRI 222
|
|
| GsiA |
COG1123 |
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain ... |
899-1115 |
4.62e-39 |
|
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 440740 [Multi-domain] Cd Length: 514 Bit Score: 154.29 E-value: 4.62e-39
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVY---RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSI 971
Cdd:COG1123 261 LEVRNLSKRYpvrGKGGVRAVDDVSLTLRRGETLGLVGESGSGKSTLARLLLGLLRPTSGSILFDGKDLtklsRRSLREL 340
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 972 RQNLGVCPQH--NVLFDMLTVEEHIWFYARL-KGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:COG1123 341 RRRVQMVFQDpySSLNPRMTVGDIIAEPLRLhGLLSRAERRERVAELLERVGLPPDLADRYPHELSGGQRQRVAIARALA 420
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS 1115
Cdd:COG1123 421 LEPKLLILDEPTSALDVSVQAQILNLLRDLQRelGLTYLFISHDLAVVRYIADRVAVMYDGRIVEDGPT 489
|
|
| PRK13537 |
PRK13537 |
nodulation factor ABC transporter ATP-binding protein NodI; |
1913-2123 |
6.28e-39 |
|
nodulation factor ABC transporter ATP-binding protein NodI;
Pssm-ID: 237420 [Multi-domain] Cd Length: 306 Bit Score: 148.41 E-value: 6.28e-39
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGY 1992
Cdd:PRK13537 7 PIDFRNVEKRY--GDKLVVDGLSFHVQRGECFGLLGPNGAGKTTTLRMLLGLTHPDAGSISLCGEPVPSRARHARQRVGV 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITELLTGREHVEFFALLRGVP----EKEVGKVGEWAirklGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:PRK13537 85 VPQFDNLDPDFTVRENLLVFGRYFGLSaaaaRALVPPLLEFA----KLENKADAKVGELSGGMKRRLTLARALVNDPDVL 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13537 161 VLDEPTTGLDPQARHLMWERLRSLLARGKTILLTTHFMEEAERLCDRLCVIEEGR 215
|
|
| ABC_putative_ATPase |
cd03269 |
ATP-binding cassette domain of an uncharacterized transporter; This subgroup is related to the ... |
1914-2128 |
1.61e-38 |
|
ATP-binding cassette domain of an uncharacterized transporter; This subgroup is related to the subfamily A transporters involved in drug resistance, nodulation, lipid transport, and bacteriocin and lantibiotic immunity. In eubacteria and archaea, the typical organization consists of one ABC and one or two integral membranes. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region in addition to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213236 [Multi-domain] Cd Length: 210 Bit Score: 143.96 E-value: 1.61e-38
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSnihEVHQNMGYC 1993
Cdd:cd03269 1 LEVENVTKRFGRVT--ALDDISFSVEKGEIFGLLGPNGAGKTTTIRMILGIILPDSGEVLFDGKPLDI---AARNRIGYL 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGK-VGEWaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDE 2072
Cdd:cd03269 76 PEERGLYPKMKVIDQLVYLAQLKGLKKEEARRrIDEW-LERLELSEYANKRVEELSKGNQQKVQFIAAVIHDPELLILDE 154
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2073 PTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:cd03269 155 PFSGLDPVNVELLKDVIRELARAGKTVILSTHQMELVEELCDRVLLLNKGRAVLYG 210
|
|
| CcmA |
COG4133 |
ABC-type transport system involved in cytochrome c biogenesis, ATPase component ... |
1914-2104 |
2.09e-38 |
|
ABC-type transport system involved in cytochrome c biogenesis, ATPase component [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 443308 [Multi-domain] Cd Length: 206 Bit Score: 143.39 E-value: 2.09e-38
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKiyRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:COG4133 3 LEAENLSC--RRGERLLFSGLSFTLAAGEALALTGPNGSGKTTLLRILAGLLPPSAGEVLWNGEPIRDAREDYRRRLAYL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEvGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:COG4133 81 GHADGLKPELTVRENLRFWAALYGLRADR-EAIDE-ALEAVGLAGLADLPVRQLSAGQKRRVALARLLLSPAPLWLLDEP 158
|
170 180 190
....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:COG4133 159 FTALDAAGVALLAELIAAHLARGGAVLLTTH 189
|
|
| ABC_NikE_OppD_transporters |
cd03257 |
ATP-binding cassette domain of nickel/oligopeptides specific transporters; The ABC transporter ... |
899-1109 |
1.78e-37 |
|
ATP-binding cassette domain of nickel/oligopeptides specific transporters; The ABC transporter subfamily specific for the transport of dipeptides, oligopeptides (OppD), and nickel (NikDE). The NikABCDE system of E. coli belongs to this family and is composed of the periplasmic binding protein NikA, two integral membrane components (NikB and NikC), and two ATPase (NikD and NikE). The NikABCDE transporter is synthesized under anaerobic conditions to meet the increased demand for nickel resulting from hydrogenase synthesis. The molecular mechanism of nickel uptake in many bacteria and most archaea is not known. Many other members of this ABC family are also involved in the uptake of dipeptides and oligopeptides. The oligopeptide transport system (Opp) is a five-component ABC transport composed of a membrane-anchored substrate binding proteins (SRP), OppA, two transmembrane proteins, OppB and OppC, and two ATP-binding domains, OppD and OppF.
Pssm-ID: 213224 [Multi-domain] Cd Length: 228 Bit Score: 141.49 E-value: 1.78e-37
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVY--RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:cd03257 2 LEVKNLSVSFptGGGSVKALDDVSFSIKKGETLGLVGESGSGKSTLARAILGLLKPTSGSIIFDGKDLlklsRRLRKIRR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVCPQH--NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALD--VGLPPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:cd03257 82 KEIQMVFQDpmSSLNPRMTIGEQIAEPLRIHGKLSKKEARKEAVLLLLvgVGLPEEVLNRYPHELSGGQRQRVAIARALA 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03257 162 LNPKLLIADEPTSALDVSVQAQILDLLkkLQEELGLTLLFITHDLGVVAKIADRVAVMYAGKI 224
|
|
| PRK13536 |
PRK13536 |
nodulation factor ABC transporter ATP-binding protein NodI; |
1914-2153 |
4.85e-37 |
|
nodulation factor ABC transporter ATP-binding protein NodI;
Pssm-ID: 237419 [Multi-domain] Cd Length: 340 Bit Score: 144.20 E-value: 4.85e-37
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKrkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:PRK13536 42 IDLAGVSKSYGDK--AVVNGLSFTVASGECFGLLGPNGAGKSTIARMILGMTSPDAGKITVLGVPVPARARLARARIGVV 119
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:PRK13536 120 PQFDNLDLEFTVRENLLVFGRYFGMSTREIEAVIPSLLEFARLESKADARVSDLSGGMKRRLTLARALINDPQLLILDEP 199
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL-KNRFGdgyTIVVRIAGSNP 2152
Cdd:PRK13536 200 TTGLDPHARHLIWERLRSLLARGKTILLTTHFMEEAERLCDRLCVLEAGRKIAEGRPHALiDEHIG---CQVIEIYGGDP 276
|
.
gi 568925948 2153 D 2153
Cdd:PRK13536 277 H 277
|
|
| ABCC_MRP_Like |
cd03228 |
ATP-binding cassette domain of multidrug resistance protein-like transporters; The MRP ... |
899-1108 |
9.16e-37 |
|
ATP-binding cassette domain of multidrug resistance protein-like transporters; The MRP (Multidrug Resistance Protein)-like transporters are involved in drug, peptide, and lipid export. They belong to the subfamily C of the ATP-binding cassette (ABC) superfamily of transport proteins. The ABCC subfamily contains transporters with a diverse functional spectrum that includes ion transport, cell surface receptor, and toxin secretion activities. The MRP-like family, similar to all ABC proteins, have a common four-domain core structure constituted by two membrane-spanning domains, each composed of six transmembrane (TM) helices, and two nucleotide-binding domains (NBD). ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213195 [Multi-domain] Cd Length: 171 Bit Score: 137.51 E-value: 9.16e-37
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:cd03228 1 IEFKNVSFSYPGRPKPVLKDVSLTIKPGEKVAIVGPSGSGKSTLLKLLLRLYDPTSGEILIDGVDLRDlDLESLRKNIAY 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHIwfyarlkglsekhvkaemeqmaldvglppsklksktsqLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:cd03228 81 VPQDPFLFSG-TIRENI--------------------------------------LSGGQRQRIAIARALLRDPPILILD 121
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGK 1108
Cdd:cd03228 122 EATSALDPETEALILEALRALAKGKTVIVIAHRLSTIR-DADRIIVLDDGR 171
|
|
| ABC_Mj1267_LivG_branched |
cd03219 |
ATP-binding cassette component of branched chain amino acids transport system; The Mj1267/LivG ... |
900-1114 |
1.57e-36 |
|
ATP-binding cassette component of branched chain amino acids transport system; The Mj1267/LivG ABC transporter subfamily is involved in the transport of the hydrophobic amino acids leucine, isoleucine and valine. MJ1267 is a branched-chain amino acid transporter with 29% similarity to both the LivF and LivG components of the E. coli branched-chain amino acid transporter. MJ1267 contains an insertion from residues 114 to 123 characteristic of LivG (Leucine-Isoleucine-Valine) homologs. The branched-chain amino acid transporter from E. coli comprises a heterodimer of ABCs (LivF and LivG), a heterodimer of six-helix TM domains (LivM and LivH), and one of two alternative soluble periplasmic substrate binding proteins (LivK or LivJ).
Pssm-ID: 213186 [Multi-domain] Cd Length: 236 Bit Score: 139.11 E-value: 1.57e-36
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCP 979
Cdd:cd03219 2 EVRGLTKRF-GGLV-ALDDVSFSVRPGEIHGLIGPNGAGKTTLFNLISGFLRPTSGSVLFDGEDITGLPPHEIARLGIGR 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 980 --QHNVLFDMLTVEE------------HIWFYARLKGLSEKHVKAEmeqMALD-VGLPPsKLKSKTSQLSGGMQRKLSVA 1044
Cdd:cd03219 80 tfQIPRLFPELTVLEnvmvaaqartgsGLLLARARREEREARERAE---ELLErVGLAD-LADRPAGELSYGQQRRLEIA 155
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:cd03219 156 RALATDPKLLLLDEPAAGLNPEETEELAELIRELRErGITVLLVEHDMDVVMSLADRVTVLDQGRVIAEGT 226
|
|
| ABC_MetN_methionine_transporter |
cd03258 |
ATP-binding cassette domain of methionine transporter; MetN (also known as YusC) is an ... |
899-1109 |
3.53e-36 |
|
ATP-binding cassette domain of methionine transporter; MetN (also known as YusC) is an ABC-type transporter encoded by metN of the metNPQ operon in Bacillus subtilis that is involved in methionine transport. Other members of this system include the MetP permease and the MetQ substrate binding protein. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213225 [Multi-domain] Cd Length: 233 Bit Score: 138.10 E-value: 3.53e-36
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:cd03258 2 IELKNVSKVFGDTGGkvTALKDVSLSVPKGEIFGIIGRSGAGKSTLIRCINGLERPTSGSVLVDGTDLtllsGKELRKAR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:cd03258 82 RRIGMIFQHFNLLSSRTVFENVALPLEIAGVPKAEIEERVLELLELVGL-EDKADAYPAQLSGGQKQRVGIARALANNPK 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03258 161 VLLCDEATSALDPETTQSILALLRDINRelGLTIVLITHEMEVVKRICDRVAVMEKGEV 219
|
|
| FtsE |
COG2884 |
Cell division ATPase FtsE [Cell cycle control, cell division, chromosome partitioning]; |
899-1109 |
3.86e-36 |
|
Cell division ATPase FtsE [Cell cycle control, cell division, chromosome partitioning];
Pssm-ID: 442130 [Multi-domain] Cd Length: 223 Bit Score: 137.49 E-value: 3.86e-36
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGmKVAVDGLALNFYEGQITsFL-GHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQ 973
Cdd:COG2884 2 IRFENVSKRYPGG-REALSDVSLEIEKGEFV-FLtGPSGAGKSTLLKLLYGEERPTSGQVLVNGQDLsrlkRREIPYLRR 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 NLGVCPQ-HNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQmALD-VGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:COG2884 80 RIGVVFQdFRLLPDR-TVYENVALPLRVTGKSRKEIRRRVRE-VLDlVGLS-DKAKALPHELSGGEQQRVAIARALVNRP 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG2884 157 ELLLADEPTGNLDPETSWEIMELLEEInRRGTTVLIATHDLELVDRMPKRVLELEDGRL 215
|
|
| ABC_cobalt_CbiO_domain1 |
cd03225 |
First domain of the ATP-binding cassette component of cobalt transport system; Domain I of the ... |
1915-2123 |
4.30e-36 |
|
First domain of the ATP-binding cassette component of cobalt transport system; Domain I of the ABC component of a cobalt transport family found in bacteria, archaea, and eukaryota. The transition metal cobalt is an essential component of many enzymes and must be transported into cells in appropriate amounts when needed. This ABC transport system of the CbiMNQO family is involved in cobalt transport in association with the cobalamin (vitamin B12) biosynthetic pathways. Most of cobalt (Cbi) transport systems possess a separate CbiN component, the cobalt-binding periplasmic protein, and they are encoded by the conserved gene cluster cbiMNQO. Both the CbiM and CbiQ proteins are integral cytoplasmic membrane proteins, and the CbiO protein has the linker peptide and the Walker A and B motifs commonly found in the ATPase components of the ABC-type transport systems.
Pssm-ID: 213192 [Multi-domain] Cd Length: 211 Bit Score: 136.83 E-value: 4.30e-36
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYC 1993
Cdd:cd03225 1 ELKNLSFSYPDGARPALDDISLTIKKGEFVLIVGPNGSGKSTLLRLLNGLLGPTSGEVLVDGKDLTKlSLKELRRKVGLV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQfDAITELL--TGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:cd03225 81 FQ-NPDDQFFgpTVEEEVAFGLENLGLPEEEIEERVEEALELVGLEGLRDRSPFTLSGGQKQRVAIAGVLAMDPDILLLD 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2072 EPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03225 160 EPTAGLDPAGRRELLELLKKLKAEGKTIIIVTHDLDLLLELADRVIVLEDGK 211
|
|
| ABC_Class3 |
cd03229 |
ATP-binding cassette domain of the binding protein-dependent transport systems; This class is ... |
901-1108 |
1.95e-35 |
|
ATP-binding cassette domain of the binding protein-dependent transport systems; This class is comprised of all BPD (Binding Protein Dependent) systems that are largely represented in archaea and eubacteria and are primarily involved in scavenging solutes from the environment. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213196 [Multi-domain] Cd Length: 178 Bit Score: 133.85 E-value: 1.95e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSI---RQNLGV 977
Cdd:cd03229 3 LKNVSKRY--GQKTVLNDVSLNIEAGEIVALLGPSGSGKSTLLRCIAGLEEPDSGSILIDGEDLTDLEDELpplRRRIGM 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIwfyarlkglsekhvkaemeqmaldvGLPpsklksktsqLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:cd03229 81 VFQDFALFPHLTVLENI-------------------------ALG----------LSGGQQQRVALARALAMDPDVLLLD 125
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1058 EPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd03229 126 EPTSALDPITRREVRALLksLQAQLGITVVLVTHDLDEAARLADRVVVLRDGK 178
|
|
| EcfA2 |
COG1122 |
Energy-coupling factor transporter ATP-binding protein EcfA2 [Inorganic ion transport and ... |
1914-2123 |
2.18e-35 |
|
Energy-coupling factor transporter ATP-binding protein EcfA2 [Inorganic ion transport and metabolism, General function prediction only];
Pssm-ID: 440739 [Multi-domain] Cd Length: 230 Bit Score: 135.54 E-value: 2.18e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL-SNIHEVHQNMGY 1992
Cdd:COG1122 1 IELENLSFSYPGGT-PALDDVSLSIEKGEFVAIIGPNGSGKSTLLRLLNGLLKPTSGEVLVDGKDITkKNLRELRRKVGL 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQF--DAITELlTGREHVEFfALL-RGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:COG1122 80 VFQNpdDQLFAP-TVEEDVAF-GPEnLGLPREEIRERVEEALELVGLEHLADRPPHELSGGQKQRVAIAGVLAMEPEVLV 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2070 LDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG1122 158 LDEPTAGLDPRGRRELLELLKRLNKEGKTVIIVTHDLDLVAELADRVIVLDDGR 211
|
|
| ABC2_perm_RbbA |
NF033858 |
ribosome-associated ATPase/putative transporter RbbA; |
914-1108 |
2.63e-35 |
|
ribosome-associated ATPase/putative transporter RbbA;
Pssm-ID: 468210 [Multi-domain] Cd Length: 907 Bit Score: 147.19 E-value: 2.63e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 914 VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDMLTVEEH 993
Cdd:NF033858 280 TAVDHVSFRIRRGEIFGFLGSNGCGKSTTMKMLTGLLPASEGEAWLFGQPVDAGDIATRRRVGYMSQAFSLYGELTVRQN 359
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 994 IWFYARLKGLSEKHVKAEMEQMALDVGLPPSkLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWE 1073
Cdd:NF033858 360 LELHARLFHLPAAEIAARVAEMLERFDLADV-ADALPDSLPLGIRQRLSLAVAVIHKPELLILDEPTSGVDPVARDMFWR 438
|
170 180 190
....*....|....*....|....*....|....*..
gi 568925948 1074 LL--LKYRQGRTIILSTHHMDEADiLGDRIAIISHGK 1108
Cdd:NF033858 439 LLieLSREDGVTIFISTHFMNEAE-RCDRISLMHAGR 474
|
|
| ABC2_membrane_3 |
pfam12698 |
ABC-2 family transporter protein; This family is related to the ABC-2 membrane transporter ... |
1545-1871 |
3.01e-35 |
|
ABC-2 family transporter protein; This family is related to the ABC-2 membrane transporter family pfam01061.
Pssm-ID: 463674 [Multi-domain] Cd Length: 345 Bit Score: 139.06 E-value: 3.01e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1545 QALPPSHEVNDAIK--QMKKLLKLTKDSSADRFLSSLGRFMAGLDTKNN--VKVWFNNKGWHAISSFLNVINNAILRANL 1620
Cdd:pfam12698 50 RALEASPTVNLVQYvdSEEEAKEALKNGKIDGLLVIPKGFSKDLLKGESatVTVYINSSNLLVSKLILNALQSLLQQLNA 129
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1621 QKGENPSQYGITAFNHPLNLTKQQLSEvalmTTSVDVLVSICVIFAMSFVPASFVVFLIQERVSKAKHLQFISGVKPVIY 1700
Cdd:pfam12698 130 SALVLLLEALSTSAPIPVESTPLFNPQ----SGYAYYLVGLILMIIILIGAAIIAVSIVEEKESRIKERLLVSGVSPLQY 205
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1701 WLSNFVWDMCNYVVPatLVIIIFICFQQksYVSSTNLPVLALLLLLYGWSITPLMYPASFVFKIPSTAYVVLTSVNLFIG 1780
Cdd:pfam12698 206 WLGKILGDFLVGLLQ--LLIILLLLFGI--GIPFGNLGLLLLLFLLYGLAYIALGYLLGSLFKNSEDAQSIIGIVILLLS 281
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1781 InGSVATFVlelftnnkLNDINDILKSVFLIFPHFCLGRGLIDMVKNQAMADalerfgenrfvsplswdlVGRNLFAMAV 1860
Cdd:pfam12698 282 G-FFGGLFP--------LEDPPSFLQWIFSIIPFFSPIDGLLRLIYGDSLWE------------------IAPSLIILLL 334
|
330
....*....|.
gi 568925948 1861 EGVVFFLITVL 1871
Cdd:pfam12698 335 FAVVLLLLALL 345
|
|
| ABC_Metallic_Cations |
cd03235 |
ATP-binding cassette domain of the metal-type transporters; This family includes transporters ... |
900-1110 |
4.36e-35 |
|
ATP-binding cassette domain of the metal-type transporters; This family includes transporters involved in the uptake of various metallic cations such as iron, manganese, and zinc. The ATPases of this group of transporters are very similar to members of iron-siderophore uptake family suggesting that they share a common ancestor. The best characterized metal-type ABC transporters are the YfeABCD system of Y. pestis, the SitABCD system of Salmonella enterica serovar Typhimurium, and the SitABCD transporter of Shigella flexneri. Moreover other uncharacterized homologs of these metal-type transporters are mainly found in pathogens like Haemophilus or enteroinvasive E. coli isolates.
Pssm-ID: 213202 [Multi-domain] Cd Length: 213 Bit Score: 134.20 E-value: 4.36e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIrqnlGVCP 979
Cdd:cd03235 1 EVEDLTVSYGG--HPVLEDVSFEVKPGEFLAIVGPNGAGKSTLLKAILGLLKPTSGSIRVFGKPLEKERKRI----GYVP 74
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 980 QH-NVLFDM-LTVEE--------HIWFYARLKglseKHVKAEMEQmALD-VGLppSKLKSKT-SQLSGGMQRKLSVALAF 1047
Cdd:cd03235 75 QRrSIDRDFpISVRDvvlmglygHKGLFRRLS----KADKAKVDE-ALErVGL--SELADRQiGELSGGQQQRVLLARAL 147
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLC 1110
Cdd:cd03235 148 VQDPDLLLLDEPFAGVDPKTQEDIYELLRELRReGMTILVVTHDLGLVLEYFDRVLLLNRTVVA 211
|
|
| FepC |
COG1120 |
ABC-type cobalamin/Fe3+-siderophores transport system, ATPase component [Inorganic ion ... |
899-1114 |
5.20e-35 |
|
ABC-type cobalamin/Fe3+-siderophores transport system, ATPase component [Inorganic ion transport and metabolism, Coenzyme transport and metabolism];
Pssm-ID: 440737 [Multi-domain] Cd Length: 254 Bit Score: 135.17 E-value: 5.20e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSS--IRQNLG 976
Cdd:COG1120 2 LEAENLSVGYGG--RPVLDDVSLSLPPGEVTALLGPNGSGKSTLLRALAGLLKPSSGEVLLDGRDL-ASLSRreLARRIA 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEE--------HIWFYARLKGLSEKHVKAEMEQMALdvglppSKLKSKT-SQLSGGMQRKLSVALAF 1047
Cdd:COG1120 79 YVPQEPPAPFGLTVRElvalgrypHLGLFGRPSAEDREAVEEALERTGL------EHLADRPvDELSGGERQRVLIARAL 152
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:COG1120 153 AQEPPLLLLDEPTSHLDLAHQLEVLELLrrLARERGRTVVMVLHDLNLAARYADRLVLLKDGRIVAQGP 221
|
|
| FetA |
COG4619 |
ABC-type iron transporter FetAB, ATPase component [Inorganic ion transport and metabolism]; |
900-1109 |
9.40e-35 |
|
ABC-type iron transporter FetAB, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443661 [Multi-domain] Cd Length: 209 Bit Score: 133.02 E-value: 9.40e-35
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVkvYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMS--SIRQNLGV 977
Cdd:COG4619 2 ELEGLS--FRVGGKPILSPVSLTLEAGECVAITGPSGSGKSTLLRALADLDPPTSGEIYLDGKPL-SAMPppEWRRQVAY 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHI--WFYARLKGLSEKHVKAEMEQmaldVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:COG4619 79 VPQEPALWGG-TVRDNLpfPFQLRERKFDRERALELLER----LGLPPDILDKPVERLSGGERQRLALIRALLLQPDVLL 153
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1056 LDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG4619 154 LDEPTSALDPENTRRVEELLREYLAeeGRAVLWVSHDPEQIERVADRVLTLEAGRL 209
|
|
| ZnuC |
COG1121 |
ABC-type Mn2+/Zn2+ transport system, ATPase component [Inorganic ion transport and metabolism]; ... |
899-1111 |
1.26e-34 |
|
ABC-type Mn2+/Zn2+ transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 440738 [Multi-domain] Cd Length: 245 Bit Score: 134.06 E-value: 1.26e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEmssiRQNLGVC 978
Cdd:COG1121 7 IELENLTVSYGG--RPVLEDVSLTIPPGEFVAIVGPNGAGKSTLLKAILGLLPPTSGTVRLFGKPPRRA----RRRIGYV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQH-NVLFDM-LTVEE--------HIWFYARLKGLSEKHVKAEMEQmaldVGLppSKLKSKT-SQLSGGMQRKLSVALAF 1047
Cdd:COG1121 81 PQRaEVDWDFpITVRDvvlmgrygRRGLFRRPSRADREAVDEALER----VGL--EDLADRPiGELSGGQQQRVLLARAL 154
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLCC 1111
Cdd:COG1121 155 AQDPDLLLLDEPFAGVDAATEEALYELLRELRrEGKTILVVTHDLGAVREYFDRVLLLNRGLVAH 219
|
|
| ABC_ATPase |
cd00267 |
ATP-binding cassette transporter nucleotide-binding domain; ABC transporters are a large ... |
1915-2123 |
1.77e-34 |
|
ATP-binding cassette transporter nucleotide-binding domain; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213179 [Multi-domain] Cd Length: 157 Bit Score: 130.44 E-value: 1.77e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSN-IHEVHQNMGYC 1993
Cdd:cd00267 1 EIENLSFRY--GGRTALDNVSLTLKAGEIVALVGPNGSGKSTLLRAIAGLLKPTSGEILIDGKDIAKLpLEELRRRIGYV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFdaitelltgrehveffallrgvpekevgkvgewairklglvkygekyasnySGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd00267 79 PQL---------------------------------------------------SGGQRQRVALARALLLNPDLLLLDEP 107
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd00267 108 TSGLDPASRERLLELLRELAEEGRTVIIVTHDPELAELAADRVIVLKDGK 157
|
|
| ABC_PstB_phosphate_transporter |
cd03260 |
ATP-binding cassette domain of the phosphate transport system; Phosphate uptake is of ... |
899-1109 |
1.80e-34 |
|
ATP-binding cassette domain of the phosphate transport system; Phosphate uptake is of fundamental importance in the cell physiology of bacteria because phosphate is required as a nutrient. The Pst system of E. coli comprises four distinct subunits encoded by the pstS, pstA, pstB, and pstC genes. The PstS protein is a phosphate-binding protein located in the periplasmic space. PstA and PstC are hydrophobic and they form the transmembrane portion of the Pst system. PstB is the catalytic subunit, which couples the energy of ATP hydrolysis to the import of phosphate across cellular membranes through the Pst system, often referred as ABC-protein. PstB belongs to one of the largest superfamilies of proteins characterized by a highly conserved adenosine triphosphate (ATP) binding cassette (ABC), which is also a nucleotide binding domain (NBD).
Pssm-ID: 213227 [Multi-domain] Cd Length: 227 Bit Score: 133.07 E-value: 1.80e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP-----PTSGTAYILGKDIRS---EMSS 970
Cdd:cd03260 1 IELRDLNVYYGD--KHALKDISLDIPKGEITALIGPSGCGKSTLLRLLNRLNDlipgaPDEGEVLLDGKDIYDldvDVLE 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQNLGVCPQHNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALD-VGLPPS-KLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:cd03260 79 LRRRVGMVFQKPNPFPG-SIYDNVAYGLRLHGIKLKEELDERVEEALRkAALWDEvKDRLHALGLSGGQQQRLCLARALA 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03260 158 NEPEVLLLDEPTSALDPISTAKIEELIAELKKEYTIVIVTHNMQQAARVADRTAFLLNGRL 218
|
|
| cbiO |
PRK13637 |
energy-coupling factor transporter ATPase; |
899-1114 |
2.86e-34 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237455 [Multi-domain] Cd Length: 287 Bit Score: 134.40 E-value: 2.86e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGM---KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE---MSSIR 972
Cdd:PRK13637 3 IKIENLTHIYMEGTpfeKKALDNVNIEIEDGEFVGLIGHTGSGKSTLIQHLNGLLKPTSGKIIIDGVDITDKkvkLSDIR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVC---PQHNvLFDMlTVEEHIWFYARLKGLSE----KHVKAEMEQmaldVGLPPSKLKSKTS-QLSGGMQRKLSVA 1044
Cdd:PRK13637 83 KKVGLVfqyPEYQ-LFEE-TIEKDIAFGPINLGLSEeeieNRVKRAMNI----VGLDYEDYKDKSPfELSGGQKRRVAIA 156
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13637 157 GVVAMEPKILILDEPTAGLDPKGRDEILNKIkeLHKEYNMTIILVSHSMEDVAKLADRIIVMNKGKCELQGT 228
|
|
| ABC_PotA_N |
cd03300 |
ATP-binding cassette domain of the polyamine transporter; PotA is an ABC-type transporter and ... |
899-1114 |
7.56e-34 |
|
ATP-binding cassette domain of the polyamine transporter; PotA is an ABC-type transporter and the ATPase component of the spermidine/putrescine-preferential uptake system consisting of PotA, -B, -C, and -D. PotA has two domains with the N-terminal domain containing the ATPase activity and the residues required for homodimerization with PotA and heterdimerization with PotB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213267 [Multi-domain] Cd Length: 232 Bit Score: 131.21 E-value: 7.56e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:cd03300 1 IELENVSKFY--GGFVALDGVSLDIKEGEFFTLLGPSGCGKTTLLRLIAGFETPTSGEILLDGKDI-TNLPPHKRPVNTV 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03300 78 FQNYALFPHLTVFENIAFGLRLKKLPKAEIKERVAEALDLVQL-EGYANRKPSQLSGGQQQRVAIARALVNEPKVLLLDE 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1059 PTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:cd03300 157 PLGALDLKLRKDMQLELkrLQKELGITFVFVTHDQEEALTMSDRIAVMNKGKIQQIGT 214
|
|
| ABC_TM1139_LivF_branched |
cd03224 |
ATP-binding cassette domain of branched-chain amino acid transporter; LivF (TM1139) is part of ... |
900-1108 |
9.48e-34 |
|
ATP-binding cassette domain of branched-chain amino acid transporter; LivF (TM1139) is part of the LIV-I bacterial ABC-type two-component transport system that imports neutral, branched-chain amino acids. The E. coli branched-chain amino acid transporter comprises a heterodimer of ABC transporters (LivF and LivG), a heterodimer of six-helix TM domains (LivM and LivH), and one of two alternative soluble periplasmic substrate binding proteins (LivK or LivJ). ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules.
Pssm-ID: 213191 [Multi-domain] Cd Length: 222 Bit Score: 130.63 E-value: 9.48e-34
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSeMSS---IRQNLG 976
Cdd:cd03224 2 EVENLNAGY--GKSQILFGVSLTVPEGEIVALLGRNGAGKTTLLKTIMGLLPPRSGSIRFDGRDITG-LPPherARAGIG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIWFYARLkgLSEKHVKAEMEQMaldVGLPP---SKLKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:cd03224 79 YVPEGRRIFPELTVEENLLLGAYA--RRRAKRKARLERV---YELFPrlkERRKQLAGTLSGGEQQMLAIARALMSRPKL 153
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd03224 154 LLLDEPSEGLAPKIVEEIFEAIRELRdEGVTILLVEQNARFALEIADRAYVLERGR 209
|
|
| GsiA |
COG1123 |
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain ... |
899-1109 |
1.61e-33 |
|
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 440740 [Multi-domain] Cd Length: 514 Bit Score: 137.73 E-value: 1.61e-33
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPT---SGTAYILGKDIRSEMSSIR-QN 974
Cdd:COG1123 5 LEVRDLSVRYPGGDVPAVDGVSLTIAPGETVALVGESGSGKSTLALALMGLLPHGgriSGEVLLDGRDLLELSEALRgRR 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:COG1123 85 IGMVFQDpMTQLNPVTVGDQIAEALENLGLSRAEARARVLELLEAVGLE-RRLDRYPHQLSGGQRQRVAIAMALALDPDL 163
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG1123 164 LIADEPTTALDVTTQAEILDLLreLQRERGTTVLLITHDLGVVAEIADRVVVMDDGRI 221
|
|
| ABC_YhbG |
cd03218 |
ATP-binding cassette component of YhbG transport system; The ABC transporters belonging to the ... |
899-1128 |
2.46e-33 |
|
ATP-binding cassette component of YhbG transport system; The ABC transporters belonging to the YhbG family are similar to members of the Mj1267_LivG family, which is involved in the transport of branched-chain amino acids. The genes yhbG and yhbN are located in a single operon and may function together in cell envelope during biogenesis. YhbG is the putative ATP-binding cassette component and YhbN is the putative periplasmic-binding protein. Depletion of each gene product leads to growth arrest, irreversible cell damage and loss of viability in E. coli. The YhbG homolog (NtrA) is essential in Rhizobium meliloti, a symbiotic nitrogen-fixing bacterium.
Pssm-ID: 213185 [Multi-domain] Cd Length: 232 Bit Score: 129.97 E-value: 2.46e-33
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03218 1 LRAENLSKRY--GKRKVVNGVSLSVKQGEIVGLLGPNGAGKTTTFYMIVGLVKPDSGKILLDGQDITKLPMHKRARLGIG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 --PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03218 79 ylPQEASIFRKLTVEENILAVLEIRGLSKKEREEKLEELLEEFHITHLR-KSKASSLSGGERRRVEIARALATNPKFLLL 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYRQgRTI-ILST-HHMDEADILGDRIAIISHGKLCCVGSS-LFLKNQLGTGYYL 1128
Cdd:cd03218 158 DEPFAGVDPIAVQDIQKIIKILKD-RGIgVLITdHNVRETLSITDRAYIIYEGKVLAEGTPeEIAANELVRKVYL 231
|
|
| CydC |
COG4987 |
ABC-type transport system involved in cytochrome bd biosynthesis, fused ATPase and permease ... |
889-1128 |
2.70e-33 |
|
ABC-type transport system involved in cytochrome bd biosynthesis, fused ATPase and permease components [Energy production and conversion, Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 444011 [Multi-domain] Cd Length: 569 Bit Score: 137.98 E-value: 2.70e-33
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHLRLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR--S 966
Cdd:COG4987 324 EPAPAPGGPSLELEDVSFRYPGAGRPVLDGLSLTLPPGERVAIVGPSGSGKSTLLALLLRFLDPQSGSITLGGVDLRdlD 403
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 967 EmSSIRQNLGVCPQHNVLFDMlTVEEHIwfyaRL--KGLSEkhvkAEMEQMALDVGLPP----------SKLKSKTSQLS 1034
Cdd:COG4987 404 E-DDLRRRIAVVPQRPHLFDT-TLRENL----RLarPDATD----EELWAALERVGLGDwlaalpdgldTWLGEGGRRLS 473
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1035 GGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGS 1114
Cdd:COG4987 474 GGERRRLALARALLRDAPILLLDEPTEGLDAATEQALLADLLEALAGRTVLLITHRLAGLE-RMDRILVLEDGRIVEQGT 552
|
250
....*....|....
gi 568925948 1115 SLFLKNQLGTGYYL 1128
Cdd:COG4987 553 HEELLAQNGRYRQL 566
|
|
| cbiO |
PRK13632 |
cobalt transporter ATP-binding subunit; Provisional |
899-1114 |
4.26e-33 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237452 [Multi-domain] Cd Length: 271 Bit Score: 130.50 E-value: 4.26e-33
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGV 977
Cdd:PRK13632 8 IKVENVSFSYPNSENNALKNVSFEINEGEYVAILGHNGSGKSTISKILTGLLKPQSGEIKIDGITISKEnLKEIRKKIGI 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:PRK13632 88 IFQNpDNQFIGATVEDDIAFGLENKKVPPKKMKDIIDDLAKKVGM-EDYLDKEPQNLSGGQKQRVAIASVLALNPEIIIF 166
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYRQGR--TIILSTHHMDEAdILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13632 167 DESTSMLDPKGKREIKKIMVDLRKTRkkTLISITHDMDEA-ILADKVIVFSEGKLIAQGK 225
|
|
| ABC_Pro_Gly_Betaine |
cd03294 |
ATP-binding cassette domain of the osmoprotectant proline/glycine betaine uptake system; This ... |
895-1113 |
7.17e-33 |
|
ATP-binding cassette domain of the osmoprotectant proline/glycine betaine uptake system; This family comprises the glycine betaine/L-proline ATP binding subunit in bacteria and its equivalents in archaea. This transport system belong to the larger ATP-Binding Cassette (ABC) transporter superfamily. The characteristic feature of these transporters is the obligatory coupling of ATP hydrolysis to substrate translocation. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213261 [Multi-domain] Cd Length: 269 Bit Score: 129.69 E-value: 7.17e-33
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 895 LRLGVSIQNLVKvyRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSS 970
Cdd:cd03294 21 LAKGKSKEEILK--KTGQTVGVNDVSLDVREGEIFVIMGLSGSGKSTLLRCINRLIEPTSGKVLIDGQDIaamsRKELRE 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQN-LGVCPQHNVLFDMLTVEEHIWFYARLKGLS--EKHVKAeMEQMALdVGLPPSKlKSKTSQLSGGMQRKLSVALAF 1047
Cdd:cd03294 99 LRRKkISMVFQSFALLPHRTVLENVAFGLEVQGVPraEREERA-AEALEL-VGLEGWE-HKYPDELSGGMQQRVGLARAL 175
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03294 176 AVDPDILLMDEAFSALDPLIRREMQDELLRLqaELQKTIVFITHDLDEALRLGDRIAIMKDGRLVQVG 243
|
|
| LivG |
COG0411 |
ABC-type branched-chain amino acid transport system, ATPase component LivG [Amino acid ... |
914-1109 |
2.44e-32 |
|
ABC-type branched-chain amino acid transport system, ATPase component LivG [Amino acid transport and metabolism];
Pssm-ID: 440180 [Multi-domain] Cd Length: 257 Bit Score: 127.85 E-value: 2.44e-32
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 914 VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCP--QHNVLFDMLTVE 991
Cdd:COG0411 18 VAVDDVSLEVERGEIVGLIGPNGAGKTTLFNLITGFYRPTSGRILFDGRDITGLPPHRIARLGIARtfQNPRLFPELTVL 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 992 EHI-----------WFYARLKGLSEKHVKAEMEQMALD----VGLPPsKLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:COG0411 98 ENVlvaaharlgrgLLAALLRLPRARREEREARERAEEllerVGLAD-RADEPAGNLSYGQQRRLEIARALATEPKLLLL 176
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYR--QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG0411 177 DEPAAGLNPEETEELAELIRRLRdeRGITILLIEHDMDLVMGLADRIVVLDFGRV 231
|
|
| ABC_NatA_like |
cd03267 |
ATP-binding cassette domain of an uncharacterized transporter similar in sequence to NatA; ... |
897-1109 |
4.56e-32 |
|
ATP-binding cassette domain of an uncharacterized transporter similar in sequence to NatA; NatA is the ATPase component of a bacterial ABC-type Na+ transport system called NatAB, which catalyzes ATP-dependent electrogenic Na+ extrusion without mechanically coupled to proton or K+ uptake. NatB possess six putative membrane spanning regions at its C-terminus. In B. subtilis, NatAB is inducible by agents such as ethanol and protonophores, which lower the proton-motive force across the membrane. The closest sequence similarity to NatA is exhibited by DrrA of the two-component daunorubicin- and doxorubicin-efflux system. Hence, the functional NatAB is presumably assembled with two copies of the single ATP-binding protein and the single integral membrane protein.
Pssm-ID: 213234 [Multi-domain] Cd Length: 236 Bit Score: 126.29 E-value: 4.56e-32
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVK-VYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNL 975
Cdd:cd03267 19 LIGSLKSLFKrKYRE--VEALKGISFTIEKGEIVGFIGPNGAGKTTTLKILSGLLQPTSGEVRVAGLVPWKRRKKFLRRI 96
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 G-VCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMA--LDVGlppSKLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:cd03267 97 GvVFGQKTQLWWDLPVIDSFYLLAAIYDLPPARFKKRLDELSelLDLE---ELLDTPVRQLSLGQRMRAEIAAALLHEPE 173
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKYRQGR--TIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03267 174 ILFLDEPTIGLDVVAQENIRNFLKEYNRERgtTVLLTSHYMKDIEALARRVLVIDKGRL 232
|
|
| ABC_ModC_like |
cd03299 |
ATP-binding cassette domain similar to the molybdate transporter; Archaeal protein closely ... |
901-1113 |
1.44e-31 |
|
ATP-binding cassette domain similar to the molybdate transporter; Archaeal protein closely related to ModC. ModC is an ABC-type transporter and the ATPase component of a molybdate transport system that also includes the periplasmic binding protein ModA and the membrane protein ModB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213266 [Multi-domain] Cd Length: 235 Bit Score: 124.76 E-value: 1.44e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYRDgmkVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVCPQ 980
Cdd:cd03299 3 VENLSKDWKE---FKLKNVSLEVERGDYFVILGPTGSGKSVLLETIAGFIKPDSGKILLNGKDI-TNLPPEKRDISYVPQ 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPT 1060
Cdd:cd03299 79 NYALFPHMTVYKNIAYGLKKRKVDKKEIERKVLEIAEMLGID-HLLNRKPETLSGGEQQRVAIARALVVNPKILLLDEPF 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1061 AGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03299 158 SALDVRTKEKLREELKKIRKefGVTVLHVTHDFEEAWALADKVAIMLNGKLIQVG 212
|
|
| ABC_PhnC_transporter |
cd03256 |
ATP-binding cassette domain of the binding protein-dependent phosphonate transport system; ... |
901-1108 |
1.76e-31 |
|
ATP-binding cassette domain of the binding protein-dependent phosphonate transport system; Phosphonates are a class of organophosphorus compounds characterized by a chemically stable carbon-to-phosphorus (C-P) bond. Phosphonates are widespread among naturally occurring compounds in all kingdoms of wildlife, but only prokaryotic microorganisms are able to cleave this bond. Certain bacteria such as E. coli can use alkylphosphonates as a phosphorus source. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213223 [Multi-domain] Cd Length: 241 Bit Score: 124.60 E-value: 1.76e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQNLG 976
Cdd:cd03256 3 VENLSKTYPNG-KKALKDVSLSINPGEFVALIGPSGAGKSTLLRCLNGLVEPTSGSVLIDGTDInklkGKALRQLRRQIG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHI---------WFYARLKGLSEKHVKAEMEqmALD-VGLpPSKLKSKTSQLSGGMQRKLSVALA 1046
Cdd:cd03256 82 MIFQQFNLIERLSVLENVlsgrlgrrsTWRSLFGLFPKEEKQRALA--ALErVGL-LDKAYQRADQLSGGQQQRVAIARA 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1047 FVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:cd03256 159 LMQQPKLILADEPVASLDPASSRQVMDLLKRINReeGITVIVSLHQVDLAREYADRIVGLKDGR 222
|
|
| COG4586 |
COG4586 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
1913-2218 |
2.59e-31 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 443643 [Multi-domain] Cd Length: 323 Bit Score: 126.74 E-value: 2.59e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP-------------------AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGD-TPvTRGDA 1972
Cdd:COG4586 1 IIEVENLSKTYRVYEKEpglkgalkglfrreyreveAVDDISFTIEPGEIVGFIGPNGAGKSTTIKMLTGIlVP-TSGEV 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1973 -------FLNKNSILSNIHEVhqnMGycpQ-----FDaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLvkyG 2040
Cdd:COG4586 80 rvlgyvpFKRRKEFARRIGVV---FG---QrsqlwWD-----LPAIDSFRLLKAIYRIPDAEYKKRLDELVELLDL---G 145
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2041 EKYasnysggNK--RKLS--------TAMALIGGPPVVFLDEPTTGMDPKA----RRFLwncaLSIVKE-GRSVVLTSHS 2105
Cdd:COG4586 146 ELL-------DTpvRQLSlgqrmrceLAAALLHRPKILFLDEPTIGLDVVSkeaiREFL----KEYNRErGTTILLTSHD 214
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2106 MEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIVVRIAGSNPDLKpvqeffgLAFPGSVLK-EKHRnmLQYQLPS 2184
Cdd:COG4586 215 MDDIEALCDRVIVIDHGRIIYDGSLEELKERFGPYKTIVLELAEPVPPLE-------LPRGGEVIErEGNR--VRLEVDP 285
|
330 340 350
....*....|....*....|....*....|....
gi 568925948 2185 SLsSLARIFSILSQskkRLHIEDYSVSQTTLDQV 2218
Cdd:COG4586 286 RE-SLAEVLARLLA---RYPVRDLTIEEPPIEEV 315
|
|
| CydD |
COG4988 |
ABC-type transport system involved in cytochrome bd biosynthesis, ATPase and permease ... |
889-1114 |
7.07e-31 |
|
ABC-type transport system involved in cytochrome bd biosynthesis, ATPase and permease components [Energy production and conversion, Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 444012 [Multi-domain] Cd Length: 563 Bit Score: 130.65 E-value: 7.07e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHLRLGVSIQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-E 967
Cdd:COG4988 327 APLPAAGPPSIELEDVSFSYPGG-RPALDGLSLTIPPGERVALVGPSGAGKSTLLNLLLGFLPPYSGSILINGVDLSDlD 405
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MSSIRQNLGVCPQHNVLFDMlTVEEHIWFYARlkGLSEkhvkAEMEQMALDVGLP------PSKLKSKT----SQLSGG- 1036
Cdd:COG4988 406 PASWRRQIAWVPQNPYLFAG-TIRENLRLGRP--DASD----EELEAALEAAGLDefvaalPDGLDTPLgeggRGLSGGq 478
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1037 MQRkLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGS 1114
Cdd:COG4988 479 AQR-LALARALLRDAPLLLLDEPTAHLDAETEAEILQALRRLAKGRTVILITHRLALLA-QADRILVLDDGRIVEQGT 554
|
|
| COG4586 |
COG4586 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
899-1109 |
9.48e-31 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 443643 [Multi-domain] Cd Length: 323 Bit Score: 125.20 E-value: 9.48e-31
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYR-----DGMK--------------VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYI 959
Cdd:COG4586 2 IEVENLSKTYRvyekePGLKgalkglfrreyrevEAVDDISFTIEPGEIVGFIGPNGAGKSTTIKMLTGILVPTSGEVRV 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 960 LGKDIRSEMSSIRQNLGVcpqhnV-------LFDmLTVEEHIWFYARLKGLSEKHVKAEMEQMA--LDVGlppSKLKSKT 1030
Cdd:COG4586 82 LGYVPFKRRKEFARRIGV-----VfgqrsqlWWD-LPAIDSFRLLKAIYRIPDAEYKKRLDELVelLDLG---ELLDTPV 152
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1031 SQLSGGmQR-KLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDeaDI--LGDRIAIIS 1105
Cdd:COG4586 153 RQLSLG-QRmRCELAAALLHRPKILFLDEPTIGLDVVSKEAIREFLKEYNRerGTTILLTSHDMD--DIeaLCDRVIVID 229
|
....
gi 568925948 1106 HGKL 1109
Cdd:COG4586 230 HGRI 233
|
|
| LPS_export_lptB |
TIGR04406 |
LPS export ABC transporter ATP-binding protein; Members of this fmaily are LptB, the ... |
900-1115 |
1.47e-30 |
|
LPS export ABC transporter ATP-binding protein; Members of this fmaily are LptB, the ATP-binding cassette protein of an ABC transporter involved in lipopolysaccharide export. [Cell envelope, Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides, Transport and binding proteins, Other]
Pssm-ID: 275199 [Multi-domain] Cd Length: 239 Bit Score: 122.00 E-value: 1.47e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGV-- 977
Cdd:TIGR04406 3 VAENLIKSY--KKRKVVNDVSLSVKSGEIVGLLGPNGAGKTTSFYMIVGLVRPDAGKILIDGQDITHLPMHERARLGIgy 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIwfYARL---KGLSEKHVKAEMEQMALDVGLppSKLKSKTSQ-LSGGMQRKLSVALAFVGGSKV 1053
Cdd:TIGR04406 81 LPQEASIFRKLTVEENI--MAVLeirKDLDRAEREERLEALLEEFQI--SHLRDNKAMsLSGGERRRVEIARALATNPKF 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS 1115
Cdd:TIGR04406 157 ILLDEPFAGVDPIAVGDIKKIIKHLKErGIGVLITDHNVRETLDICDRAYIISDGKVLAEGTP 219
|
|
| ABC_Carb_Solutes_like |
cd03259 |
ATP-binding cassette domain of the carbohydrate and solute transporters-like; This family is ... |
1914-2123 |
2.51e-30 |
|
ATP-binding cassette domain of the carbohydrate and solute transporters-like; This family is comprised of proteins involved in the transport of apparently unrelated solutes and proteins specific for di- and oligosaccharides and polyols. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213226 [Multi-domain] Cd Length: 213 Bit Score: 120.32 E-value: 2.51e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNihEVHQ-NMGY 1992
Cdd:cd03259 1 LELKGLSKTYGSVR--ALDDLSLTVEPGEFLALLGPSGCGKTTLLRLIAGLERPDSGEILIDGRDVTGV--PPERrNIGM 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITELLTGREHVEFfAL-LRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:cd03259 77 VFQDYALFPHLTVAENIAF-GLkLRGVPKAEIRARVRELLELVGLEGLLNRYPHELSGGQQQRVALARALAREPSLLLLD 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2072 EPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03259 156 EPLSALDAKLREELREELKELQRElGITTIYVTHDQEEALALADRIAVMNEGR 208
|
|
| NupO |
COG3845 |
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and ... |
899-1114 |
3.33e-30 |
|
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and metabolism];
Pssm-ID: 443055 [Multi-domain] Cd Length: 504 Bit Score: 127.45 E-value: 3.33e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK--DIRSEMSSIRQNLG 976
Cdd:COG3845 6 LELRGITKRF--GGVVANDDVSLTVRPGEIHALLGENGAGKSTLMKILYGLYQPDSGEILIDGKpvRIRSPRDAIALGIG 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIW--------FYARLKGLSEKhVKAEMEQMALDVglPPSKlksKTSQLSGGMQRKLSVALAFV 1048
Cdd:COG3845 84 MVHQHFMLVPNLTVAENIVlgleptkgGRLDRKAARAR-IRELSERYGLDV--DPDA---KVEDLSVGEQQRVEILKALY 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLccVGS 1114
Cdd:COG3845 158 RGARILILDEPTAVLTPQEADELFEILRRLAaEGKSIIFITHKLREVMAIADRVTVLRRGKV--VGT 222
|
|
| FepC |
COG1120 |
ABC-type cobalamin/Fe3+-siderophores transport system, ATPase component [Inorganic ion ... |
1913-2131 |
4.05e-30 |
|
ABC-type cobalamin/Fe3+-siderophores transport system, ATPase component [Inorganic ion transport and metabolism, Coenzyme transport and metabolism];
Pssm-ID: 440737 [Multi-domain] Cd Length: 254 Bit Score: 121.30 E-value: 4.05e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMG 1991
Cdd:COG1120 1 MLEAENLS--VGYGGRPVLDDVSLSLPPGEVTALLGPNGSGKSTLLRALAGLLKPSSGEVLLDGRDLASlSRRELARRIA 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDAITELLTGREHVE-----FFALLRGVPEKEVGKVgEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPP 2066
Cdd:COG1120 79 YVPQEPPAPFGLTVRELVAlgrypHLGLFGRPSAEDREAV-EEALERTGLEHLADRPVDELSGGERQRVLIARALAQEPP 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2067 VVFLDEPTTGMDPKARRFLwncaLSIVKE-----GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQ 2131
Cdd:COG1120 158 LLLLDEPTSHLDLAHQLEV----LELLRRlarerGRTVVMVLHDLNLAARYADRLVLLKDGRIVAQGPPE 223
|
|
| ABC_Iron-Siderophores_B12_Hemin |
cd03214 |
ATP-binding component of iron-siderophores, vitamin B12 and hemin transporters and related ... |
900-1113 |
5.06e-30 |
|
ATP-binding component of iron-siderophores, vitamin B12 and hemin transporters and related proteins; ABC transporters, involved in the uptake of siderophores, heme, and vitamin B12, are widely conserved in bacteria and archaea. Only very few species lack representatives of the siderophore family transporters. The E. coli BtuCD protein is an ABC transporter mediating vitamin B12 uptake. The two ATP-binding cassettes (BtuD) are in close contact with each other, as are the two membrane-spanning subunits (BtuC); this arrangement is distinct from that observed for the E. coli lipid flippase MsbA. The BtuC subunits provide 20 transmembrane helices grouped around a translocation pathway that is closed to the cytoplasm by a gate region, whereas the dimer arrangement of the BtuD subunits resembles the ATP-bound form of the Rad50 DNA repair enzyme. A prominent cytoplasmic loop of BtuC forms the contact region with the ATP-binding cassette and represent a conserved motif among the ABC transporters.
Pssm-ID: 213181 [Multi-domain] Cd Length: 180 Bit Score: 118.31 E-value: 5.06e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSS--IRQNLGV 977
Cdd:cd03214 1 EVENLSVGYGG--RTVLDDLSLSIEAGEIVGILGPNGAGKSTLLKTLAGLLKPSSGEILLDGKDL-ASLSPkeLARKIAY 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLtveehiwfyarlkGLSEKHVkaemeqmaldvglppsklksktSQLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:cd03214 78 VPQALELLGLA-------------HLADRPF----------------------NELSGGERQRVLLARALAQEPPILLLD 122
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1058 EPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03214 123 EPTSHLDIAHQIELLELLrrLARERGKTVVMVLHDLNLAARYADRVILLKDGRIVAQG 180
|
|
| ABC_MalK_N |
cd03301 |
The N-terminal ATPase domain of the maltose transporter, MalK; ATP binding cassette (ABC) ... |
899-1113 |
9.86e-30 |
|
The N-terminal ATPase domain of the maltose transporter, MalK; ATP binding cassette (ABC) proteins function from bacteria to human, mediating the translocation of substances into and out of cells or organelles. ABC transporters contain two transmembrane-spanning domains (TMDs) or subunits and two nucleotide binding domains (NBDs) or subunits that couple transport to the hydrolysis of ATP. In the maltose transport system, the periplasmic maltose binding protein (MBP) stimulates the ATPase activity of the membrane-associated transporter, which consists of two transmembrane subunits, MalF and MalG, and two copies of the ATP binding subunit, MalK, and becomes tightly bound to the transporter in the catalytic transition state, ensuring that maltose is passed to the transporter as ATP is hydrolyzed.
Pssm-ID: 213268 [Multi-domain] Cd Length: 213 Bit Score: 118.90 E-value: 9.86e-30
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:cd03301 1 VELENVTKRF--GNVTALDDLNLDIADGEFVVLLGPSGCGKTTTLRMIAGLEEPTSGRIYIGGRDV-TDLPPKDRDIAMV 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMA--LDVGlppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03301 78 FQNYALYPHMTVYDNIAFGLKLRKVPKDEIDERVREVAelLQIE---HLLDRKPKQLSGGQRQRVALGRAIVREPKVFLM 154
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1057 DEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03301 155 DEPLSNLDAKLRVQMRAELkrLQQRLGTTTIYVTHDQVEAMTMADRIAVMNDGQIQQIG 213
|
|
| ABC_FtsE |
cd03292 |
Cell division ATP-binding protein FtsE; The FtsEX complex resembles an ABC transporter, where ... |
902-1109 |
1.24e-29 |
|
Cell division ATP-binding protein FtsE; The FtsEX complex resembles an ABC transporter, where FtsE is the ATPase and the membrane subunit FtsX resembles a permease subunit. But rather than transporting any substrate, the complex acts in cell division by undergoing conformational changes that alter the activity of cell wall hydrolases located outside the plasma membrane. The complex is widely conserved in bacteria, but also extremely divergent in sequence between different lineages
Pssm-ID: 213259 [Multi-domain] Cd Length: 214 Bit Score: 118.66 E-value: 1.24e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 902 QNLVKVYrDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQNLGV 977
Cdd:cd03292 4 INVTKTY-PNGTAALDGINISISAGEFVFLVGPSGAGKSTLLKLIYKEELPTSGTIRVNGQDVsdlrGRAIPYLRRKIGV 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIWFYARLKGLSEKHVkAEMEQMALD-VGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03292 83 VFQDFRLLPDRNVYENVAFALEVTGVPPREI-RKRVPAALElVGLS-HKHRALPAELSGGEQQRVAIARAIVNSPTILIA 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1057 DEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03292 161 DEPTGNLDPDTTWEIMNLLKKINKaGTTVVVATHAKELVDTTRHRVIALERGKL 214
|
|
| MalK |
COG3839 |
ABC-type sugar transport system, ATPase component MalK [Carbohydrate transport and metabolism]; ... |
899-1114 |
1.40e-29 |
|
ABC-type sugar transport system, ATPase component MalK [Carbohydrate transport and metabolism];
Pssm-ID: 443050 [Multi-domain] Cd Length: 352 Bit Score: 122.49 E-value: 1.40e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:COG3839 4 LELENVSKSY--GGVEALKDIDLDIEDGEFLVLLGPSGCGKSTLLRMIAGLEDPTSGEILIGGRDV-TDLPPKDRNIAMV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGG-MQRklsVAL--AFVGGSKVVI 1055
Cdd:COG3839 81 FQSYALYPHMTVYENIAFPLKLRKVPKAEIDRRVREAAELLGLED-LLDRKPKQLSGGqRQR---VALgrALVREPKVFL 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1056 LDEPTAGVDPYSR---RGiwEL--LLKyRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:COG3839 157 LDEPLSNLDAKLRvemRA--EIkrLHR-RLGTTTIYVTHDQVEAMTLADRIAVMNDGRIQQVGT 217
|
|
| ABC_Carb_Monos_I |
cd03216 |
First domain of the ATP-binding cassette component of monosaccharide transport system; This ... |
899-1109 |
1.86e-29 |
|
First domain of the ATP-binding cassette component of monosaccharide transport system; This family represents the domain I of the carbohydrate uptake proteins that transport only monosaccharides (Monos). The Carb_Monos family is involved in the uptake of monosaccharides, such as pentoses (such as xylose, arabinose, and ribose) and hexoses (such as xylose, arabinose, and ribose), that cannot be broken down to simple sugars by hydrolysis. Pentoses include xylose, arabinose, and ribose. Important hexoses include glucose, galactose, and fructose. In members of the Carb_monos family, the single hydrophobic gene product forms a homodimer while the ABC protein represents a fusion of two nucleotide-binding domains. However, it is assumed that two copies of the ABC domains are present in the assembled transporter.
Pssm-ID: 213183 [Multi-domain] Cd Length: 163 Bit Score: 115.99 E-value: 1.86e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrsEMSSIR--QNLG 976
Cdd:cd03216 1 LELRGITKRF--GGVKALDGVSLSVRRGEVHALLGENGAGKSTLMKILSGLYKPDSGEILVDGKEV--SFASPRdaRRAG 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VcpqhnvlfdmLTVeehiwfyarlkglsekhvkaemeqmaldvglppsklksktSQLSGGMQRKLSVALAFVGGSKVVIL 1056
Cdd:cd03216 77 I----------AMV----------------------------------------YQLSVGERQMVEIARALARNARLLIL 106
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1057 DEPTAGVDPYSRRGIWELL--LKyRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03216 107 DEPTAALTPAEVERLFKVIrrLR-AQGVAVIFISHRLDEVFEIADRVTVLRDGRV 160
|
|
| DppF |
COG1124 |
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid ... |
899-1109 |
2.53e-29 |
|
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid transport and metabolism, Inorganic ion transport and metabolism];
Pssm-ID: 440741 [Multi-domain] Cd Length: 248 Bit Score: 118.75 E-value: 2.53e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGM--KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-RSEMSSIRQNL 975
Cdd:COG1124 2 LEVRNLSVSYGQGGrrVPVLKDVSLEVAPGESFGLVGESGSGKSTLLRALAGLERPWSGEVTFDGRPVtRRRRKAFRRRV 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQH--NVLFDMLTVEEHIwfyAR-LKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGG-MQRklsVAL--AFVG 1049
Cdd:COG1124 82 QMVFQDpyASLHPRHTVDRIL---AEpLRIHGLPDREERIAELLEQVGLPPSFLDRYPHQLSGGqRQR---VAIarALIL 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1050 GSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG1124 156 EPELLLLDEPTSALDVSVQAEILNLLKDLREerGLTYLFVSHDLAVVAHLCDRVAVMQNGRI 217
|
|
| ABC_MJ0796_LolCDE_FtsE |
cd03255 |
ATP-binding cassette domain of the transporters involved in export of lipoprotein and ... |
1914-2123 |
3.16e-29 |
|
ATP-binding cassette domain of the transporters involved in export of lipoprotein and macrolide, and Cell division ATP-binding protein FtsE; This family is comprised of MJ0796 ATP-binding cassette, macrolide-specific ABC-type efflux carrier (MacAB), and proteins involved in cell division (FtsE), and release of lipoproteins from the cytoplasmic membrane (LolCDE). They are clustered together phylogenetically. MacAB is an exporter that confers resistance to macrolides, while the LolCDE system is not a transporter at all. The FtsEX complex resembles an ABC transporter, where FtsE is the ATPase and the membrane subunit FtsX resembles a permease subunit. But rather than transporting any substrate, the complex acts in cell division by undergoing conformational changes that alter the activity of cell wall hydrolases located outside the plasma membrane. The complex is widely conserved in bacteria, but also extremely divergent in sequence between different lineages. The LolCDE complex catalyzes the release of lipoproteins from the cytoplasmic membrane prior to their targeting to the outer membrane.
Pssm-ID: 213222 [Multi-domain] Cd Length: 218 Bit Score: 117.59 E-value: 3.16e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRR--KRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIHEV----- 1986
Cdd:cd03255 1 IELKNLSKTYGGggEKVQALKGVSLSIEKGEFVAIVGPSGSGKSTLLNILGGLDRPTSGEVRVDGTDI-SKLSEKelaaf 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 -HQNMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:cd03255 80 rRRHIGFVFQSFNLLPDLTALENVELPLLLAGVPKKERRERAEELLERVGLGDRLNHYPSELSGGQQQRVAIARALANDP 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEEcEALCTRMAIMVNGR 2123
Cdd:cd03255 160 KIILADEPTGNLDSETGKEVMELLRELNKEaGTTIVVVTHDPEL-AEYADRIIELRDGK 217
|
|
| GsiA |
COG1123 |
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain ... |
1913-2133 |
3.47e-29 |
|
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 440740 [Multi-domain] Cd Length: 514 Bit Score: 124.63 E-value: 3.47e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVT---RGDAFLNKNSIL-SNIHEVHQ 1988
Cdd:COG1123 4 LLEVRDLSVRYPGGDVPAVDGVSLTIAPGETVALVGESGSGKSTLALALMGLLPHGgriSGEVLLDGRDLLeLSEALRGR 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQfDAITEL--LTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPP 2066
Cdd:COG1123 84 RIGMVFQ-DPMTQLnpVTVGDQIAEALENLGLSRAEARARVLELLEAVGLERRLDRYPHQLSGGQRQRVAIAMALALDPD 162
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2067 VVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:COG1123 163 LLIADEPTTALDVTTQAEILDLLRELQRErGTTVLLITHDLGVVAEIADRVVVMDDGRIVEDGPPEEI 230
|
|
| LptB |
COG1137 |
ABC-type lipopolysaccharide export system, ATPase component [Cell wall/membrane/envelope ... |
900-1115 |
3.86e-29 |
|
ABC-type lipopolysaccharide export system, ATPase component [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440752 [Multi-domain] Cd Length: 240 Bit Score: 117.82 E-value: 3.86e-29
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-------RSemssiR 972
Cdd:COG1137 5 EAENLVKSYGK--RTVVKDVSLEVNQGEIVGLLGPNGAGKTTTFYMIVGLVKPDSGRIFLDGEDIthlpmhkRA-----R 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLppSKL-KSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:COG1137 78 LGIGYLPQEASIFRKLTVEDNILAVLELRKLSKKEREERLEELLEEFGI--THLrKSKAYSLSGGERRRVEIARALATNP 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYRQgRTI-ILST-HHMDEadILG--DRIAIISHGKLCCVGSS 1115
Cdd:COG1137 156 KFILLDEPFAGVDPIAVADIQKIIRHLKE-RGIgVLITdHNVRE--TLGicDRAYIISEGKVLAEGTP 220
|
|
| ZnuC |
COG1121 |
ABC-type Mn2+/Zn2+ transport system, ATPase component [Inorganic ion transport and metabolism]; ... |
1913-2132 |
1.44e-28 |
|
ABC-type Mn2+/Zn2+ transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 440738 [Multi-domain] Cd Length: 245 Bit Score: 116.34 E-value: 1.44e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKrkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKnsilSNIHEVHQNMGY 1992
Cdd:COG1121 6 AIELENLTVSYGGR--PVLEDVSLTIPPGEFVAIVGPNGAGKSTLLKAILGLLPPTSGTVRLFG----KPPRRARRRIGY 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITEL--LTGREHVEF-----FALLRGVPEKEVGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:COG1121 80 VPQRAEVDWDfpITVRDVVLMgrygrRGLFRRPSRADREAVDE-ALERVGLEDLADRPIGELSGGQQQRVLLARALAQDP 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRmAIMVNGRFRCLGSVQH 2132
Cdd:COG1121 159 DLLLLDEPFAGVDAATEEALYELLRELRREGKTILVVTHDLGAVREYFDR-VLLLNRGLVAHGPPEE 224
|
|
| cbiO |
TIGR01166 |
cobalt transport protein ATP-binding subunit; This model describes the ATP binding subunit of ... |
908-1094 |
1.75e-28 |
|
cobalt transport protein ATP-binding subunit; This model describes the ATP binding subunit of the multisubunit cobalt transporter in bacteria and its equivalents in archaea. The model is restricted to ATP subunit that is a part of the cobalt transporter, which belongs to the ABC transporter superfamily (ATP Binding Cassette). The model excludes ATP binding subunit that are associated with other transporters belonging to ABC transporter superfamily. This superfamily includes two groups, one which catalyze the uptake of small molecules, including ions from the external milieu and the other group which is engaged in the efflux of small molecular weight compounds and ions from within the cell. Energy derived from the hydrolysis of ATP drive the both the process of uptake and efflux. [Transport and binding proteins, Cations and iron carrying compounds]
Pssm-ID: 130234 [Multi-domain] Cd Length: 190 Bit Score: 114.44 E-value: 1.75e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI---RSEMSSIRQNLGVCPQH--N 982
Cdd:TIGR01166 1 YPGGPEV-LKGLNFAAERGEVLALLGANGAGKSTLLLHLNGLLRPQSGAVLIDGEPLdysRKGLLERRQRVGLVFQDpdD 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 983 VLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLppSKLKSK-TSQLSGGMQRKLSVALAFVGGSKVVILDEPTA 1061
Cdd:TIGR01166 80 QLFAA-DVDQDVAFGPLNLGLSEAEVERRVREALTAVGA--SGLRERpTHCLSGGEKKRVAIAGAVAMRPDVLLLDEPTA 156
|
170 180 190
....*....|....*....|....*....|....
gi 568925948 1062 GVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEA 1094
Cdd:TIGR01166 157 GLDPAGREQMLAILRRLRaEGMTVVISTHDVDLA 190
|
|
| ABC_CysA_sulfate_importer |
cd03296 |
ATP-binding cassette domain of the sulfate transporter; Part of the ABC transporter complex ... |
897-1114 |
1.85e-28 |
|
ATP-binding cassette domain of the sulfate transporter; Part of the ABC transporter complex cysAWTP involved in sulfate import. Responsible for energy coupling to the transport system. The complex is composed of two ATP-binding proteins (cysA), two transmembrane proteins (cysT and cysW), and a solute-binding protein (cysP). ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213263 [Multi-domain] Cd Length: 239 Bit Score: 115.90 E-value: 1.85e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLG 976
Cdd:cd03296 1 MSIEVRNVSKRF--GDFVALDDVSLDIPSGELVALLGPSGSGKTTLLRLIAGLERPDSGTILFGGEDA-TDVPVQERNVG 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALD----VGLppSKLKSK-TSQLSGGMQRKLSVALAFVGGS 1051
Cdd:cd03296 78 FVFQHYALFRHMTVFDNVAFGLRVKPRSERPPEAEIRAKVHEllklVQL--DWLADRyPAQLSGGQRQRVALARALAVEP 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1052 KVVILDEPTAGVDPYSRRGI--WELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:cd03296 156 KVLLLDEPFGALDAKVRKELrrWLRRLHDELHVTTVFVTHDQEEALEVADRVVVMNKGRIEQVGT 220
|
|
| cbiO |
PRK13647 |
cobalt transporter ATP-binding subunit; Provisional |
899-1121 |
4.20e-28 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237457 [Multi-domain] Cd Length: 274 Bit Score: 115.99 E-value: 4.20e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS-IRQNLGV 977
Cdd:PRK13647 5 IEVEDLHFRYKDGTK-ALKGLSLSIPEGSKTALLGPNGAGKSTLLLHLNGIYLPQRGRVKVMGREVNAENEKwVRSKVGL 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQ--HNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQmALD-VGLppSKLKSKTS-QLSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK13647 84 VFQdpDDQVFSS-TVWDDVAFGPVNMGLDKDEVERRVEE-ALKaVRM--WDFRDKPPyHLSYGQKKRVAIAGVLAMDPDV 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQ 1121
Cdd:PRK13647 160 IVLDEPMAYLDPRGQETLMEILDRlHNQGKTVIVATHDVDLAAEWADQVIVLKEGRVLAEGDKSLLTDE 228
|
|
| cbiO |
PRK13641 |
energy-coupling factor transporter ATPase; |
897-1109 |
5.08e-28 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237456 [Multi-domain] Cd Length: 287 Bit Score: 116.08 E-value: 5.08e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVKVYRDGM---KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS--- 970
Cdd:PRK13641 1 MSIKFENVDYIYSPGTpmeKKGLDNISFELEEGSFVALVGHTGSGKSTLMQHFNALLKPSSGTITIAGYHITPETGNknl 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 --IRQNLGVCPQ--HNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALA 1046
Cdd:PRK13641 81 kkLRKKVSLVFQfpEAQLFEN-TVLKDVEFGPKNFGFSEDEAKEKALKWLKKVGLSEDLISKSPFELSGGQMRRVAIAGV 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1047 FVGGSKVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK13641 160 MAYEPEILCLDEPAAGLDPEGRKEMMQLFKDYqKAGHTVILVTHNMDDVAEYADDVLVLEHGKL 223
|
|
| ABC_tran |
pfam00005 |
ABC transporter; ABC transporters for a large family of proteins responsible for translocation ... |
1938-2075 |
5.32e-28 |
|
ABC transporter; ABC transporters for a large family of proteins responsible for translocation of a variety of compounds across biological membranes. ABC transporters are the largest family of proteins in many completely sequenced bacteria. ABC transporters are composed of two copies of this domain and two copies of a transmembrane domain pfam00664. These four domains may belong to a single polypeptide or belong in different polypeptide chains.
Pssm-ID: 394964 [Multi-domain] Cd Length: 150 Bit Score: 111.59 E-value: 5.32e-28
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQFDAITELLTGREHVEFFALLR 2016
Cdd:pfam00005 8 LNPGEILALVGPNGAGKSTLLKLIAGLLSPTEGTILLDGQDLTDdERKSLRKEIGYVFQDPQLFPRLTVRENLRLGLLLK 87
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2017 GVPEKEVGKVGEWAIRKLGLVKYGE----KYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTT 2075
Cdd:pfam00005 88 GLSKREKDARAEEALEKLGLGDLADrpvgERPGTLSGGQRQRVAIARALLTKPKLLLLDEPTA 150
|
|
| cbiO |
PRK13634 |
cobalt transporter ATP-binding subunit; Provisional |
908-1114 |
1.01e-27 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237454 [Multi-domain] Cd Length: 290 Bit Score: 115.50 E-value: 1.01e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVAVDGLAL-----NFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-----MSSIRQNLGV 977
Cdd:PRK13634 10 HRYQYKTPFERRALydvnvSIPSGSYVAIIGHTGSGKSTLLQHLNGLLQPTSGTVTIGERVITAGkknkkLKPLRKKVGI 89
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 C---PQHNvLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:PRK13634 90 VfqfPEHQ-LFEE-TVEKDICFGPMNFGVSEEDAKQKAREMIELVGLPEELLARSPFELSGGQMRRVAIAGVLAMEPEVL 167
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13634 168 VLDEPTAGLDPKGRKEMMEMFykLHKEKGLTTVLVTHSMEDAARYADQIVVMHKGTVFLQGT 229
|
|
| ABC_Mj1267_LivG_branched |
cd03219 |
ATP-binding cassette component of branched chain amino acids transport system; The Mj1267/LivG ... |
1914-2135 |
1.08e-27 |
|
ATP-binding cassette component of branched chain amino acids transport system; The Mj1267/LivG ABC transporter subfamily is involved in the transport of the hydrophobic amino acids leucine, isoleucine and valine. MJ1267 is a branched-chain amino acid transporter with 29% similarity to both the LivF and LivG components of the E. coli branched-chain amino acid transporter. MJ1267 contains an insertion from residues 114 to 123 characteristic of LivG (Leucine-Isoleucine-Valine) homologs. The branched-chain amino acid transporter from E. coli comprises a heterodimer of ABCs (LivF and LivG), a heterodimer of six-helix TM domains (LivM and LivH), and one of two alternative soluble periplasmic substrate binding proteins (LivK or LivJ).
Pssm-ID: 213186 [Multi-domain] Cd Length: 236 Bit Score: 113.69 E-value: 1.08e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEV------ 1986
Cdd:cd03219 1 LEVRGLTKRFGGLV--ALDDVSFSVRPGEIHGLIGPNGAGKTTLFNLISGFLRPTSGSVLFDGEDITGlPPHEIarlgig 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 --HQNMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWA---IRKLGLVKYGEKYASNYSGGNKRKLSTAMAL 2061
Cdd:cd03219 79 rtFQIPRLFPELTVLENVMVAAQARTGSGLLLARARREEREARERAeelLERVGLADLADRPAGELSYGQQRRLEIARAL 158
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2062 IGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:cd03219 159 ATDPKLLLLDEPAAGLNPEETEELAELIRELRERGITVLLVEHDMDVVMSLADRVTVLDQGRVIAEGTPDEVRN 232
|
|
| SunT |
COG2274 |
ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase ... |
891-1128 |
2.16e-27 |
|
ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase domain [Defense mechanisms];
Pssm-ID: 441875 [Multi-domain] Cd Length: 711 Bit Score: 121.09 E-value: 2.16e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 891 EPTHLRLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMS 969
Cdd:COG2274 466 SLPRLKGDIELENVSFRYPGDSPPVLDNISLTIKPGERVAIVGRSGSGKSTLLKLLLGLYEPTSGRILIDGIDLRQiDPA 545
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 970 SIRQNLGVCPQHNVLFDMlTVEEHIWFYArlKGLSEkhvkAEMEQMALDVGLP------PSKLKSK----TSQLSGGMQR 1039
Cdd:COG2274 546 SLRRQIGVVLQDVFLFSG-TIRENITLGD--PDATD----EEIIEAARLAGLHdfiealPMGYDTVvgegGSNLSGGQRQ 618
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1040 KLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTH---HMDEAdilgDRIAIISHGKLCCVGSSL 1116
Cdd:COG2274 619 RLAIARALLRNPRILILDEATSALDAETEAIILENLRRLLKGRTVIIIAHrlsTIRLA----DRIIVLDKGRIVEDGTHE 694
|
250
....*....|..
gi 568925948 1117 FLKNQLGTGYYL 1128
Cdd:COG2274 695 ELLARKGLYAEL 706
|
|
| cbiO |
PRK13649 |
energy-coupling factor transporter ATPase; |
897-1133 |
3.07e-27 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184208 [Multi-domain] Cd Length: 280 Bit Score: 113.69 E-value: 3.07e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVKVYRDGMKV---AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-----EM 968
Cdd:PRK13649 1 MGINLQNVSYTYQAGTPFegrALFDVNLTIEDGSYTAFIGHTGSGKSTIMQLLNGLHVPTQGSVRVDDTLITStsknkDI 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 SSIRQNLGVCPQ--HNVLFDMlTVEEHIWFYARLKGLS-EKHVKAEMEQMALdVGLPPSKLKSKTSQLSGGMQRKLSVAL 1045
Cdd:PRK13649 81 KQIRKKVGLVFQfpESQLFEE-TVLKDVAFGPQNFGVSqEEAEALAREKLAL-VGISESLFEKNPFELSGGQMRRVAIAG 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1046 AFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS--------SL 1116
Cdd:PRK13649 159 ILAMEPKILVLDEPTAGLDPKGRKELMTLFKKLHQsGMTIVLVTHLMDDVANYADFVYVLEKGKLVLSGKpkdifqdvDF 238
|
250 260
....*....|....*....|.
gi 568925948 1117 FLKNQLG----TGYYLTLVKK 1133
Cdd:PRK13649 239 LEEKQLGvpkiTKFAQRLADR 259
|
|
| ABC_YhbG |
cd03218 |
ATP-binding cassette component of YhbG transport system; The ABC transporters belonging to the ... |
1914-2135 |
5.20e-27 |
|
ATP-binding cassette component of YhbG transport system; The ABC transporters belonging to the YhbG family are similar to members of the Mj1267_LivG family, which is involved in the transport of branched-chain amino acids. The genes yhbG and yhbN are located in a single operon and may function together in cell envelope during biogenesis. YhbG is the putative ATP-binding cassette component and YhbN is the putative periplasmic-binding protein. Depletion of each gene product leads to growth arrest, irreversible cell damage and loss of viability in E. coli. The YhbG homolog (NtrA) is essential in Rhizobium meliloti, a symbiotic nitrogen-fixing bacterium.
Pssm-ID: 213185 [Multi-domain] Cd Length: 232 Bit Score: 111.48 E-value: 5.20e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHE-VHQNMG 1991
Cdd:cd03218 1 LRAENLSKRY--GKRKVVNGVSLSVKQGEIVGLLGPNGAGKTTTFYMIVGLVKPDSGKILLDGQDITKlPMHKrARLGIG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:cd03218 79 YLPQEASIFRKLTVEENILAVLEIRGLSKKEREEKLEELLEEFHITHLRKSKASSLSGGERRRVEIARALATNPKFLLLD 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2072 EPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:cd03218 159 EPFAGVDPIAVQDIQKIIKILKDRGIGVLITDHNVRETLSITDRAYIIYEGKVLAEGTPEEIAA 222
|
|
| cbiO |
PRK13639 |
cobalt transporter ATP-binding subunit; Provisional |
903-1109 |
5.22e-27 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184199 [Multi-domain] Cd Length: 275 Bit Score: 112.86 E-value: 5.22e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 903 NLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS---IRQNLGVCP 979
Cdd:PRK13639 6 DLKYSYPDG-TEALKGINFKAEKGEMVALLGPNGAGKSTLFLHFNGILKPTSGEVLIKGEPIKYDKKSlleVRKTVGIVF 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 980 QH--NVLFdMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGL------PPsklksktSQLSGGMQRKLSVALAFVGGS 1051
Cdd:PRK13639 85 QNpdDQLF-APTVEEDVAFGPLNLGLSKEEVEKRVKEALKAVGMegfenkPP-------HHLSGGQKKRVAIAGILAMKP 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK13639 157 EIIVLDEPTSGLDPMGASQIMKLLYDLnKEGITIIISTHDVDLVPVYADKVYVMSDGKI 215
|
|
| cbiO |
PRK13646 |
energy-coupling factor transporter ATPase; |
899-1136 |
7.78e-27 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184205 [Multi-domain] Cd Length: 286 Bit Score: 112.57 E-value: 7.78e-27
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGM---KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-----EMSS 970
Cdd:PRK13646 3 IRFDNVSYTYQKGTpyeHQAIHDVNTEFEQGKYYAIVGQTGSGKSTLIQNINALLKPTTGTVTVDDITITHktkdkYIRP 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQNLGVCPQ--HNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:PRK13646 83 VRKRIGMVFQfpESQLFED-TVEREIIFGPKNFKMNLDEVKNYAHRLLMDLGFSRDVMSQSPFQMSGGQMRKIAIVSILA 161
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS--SLFLKNQLGT 1124
Cdd:PRK13646 162 MNPDIIVLDEPTAGLDPQSKRQVMRLLksLQTDENKTIILVSHDMNEVARYADEVIVMKEGSIVSQTSpkELFKDKKKLA 241
|
250
....*....|....*..
gi 568925948 1125 GYYLTL-----VKKDVE 1136
Cdd:PRK13646 242 DWHIGLpeivqLQYDFE 258
|
|
| ABCG_White |
cd03234 |
White pigment protein homolog of ABCG transporter subfamily; The White subfamily represents ... |
918-1109 |
1.04e-26 |
|
White pigment protein homolog of ABCG transporter subfamily; The White subfamily represents ABC transporters homologous to the Drosophila white gene, which acts as a dimeric importer for eye pigment precursors. The eye pigmentation of Drosophila is developed from the synthesis and deposition in the cells of red pigments, which are synthesized from guanine, and brown pigments, which are synthesized from tryptophan. The pigment precursors are encoded by the white, brown, and scarlet genes, respectively. Evidence from genetic and biochemical studies suggest that the White and Brown proteins function as heterodimers to import guanine, while the White and Scarlet proteins function to import tryptophan. However, a recent study also suggests that White may be involved in the transport of a metabolite, such as 3-hydroxykynurenine, across intracellular membranes. Mammalian ABC transporters belonging to the White subfamily (ABCG1, ABCG5, and ABCG8) have been shown to be involved in the regulation of lipid-trafficking mechanisms in macrophages, hepatocytes, and intestinal mucosa cells. ABCG1 (ABC8), the human homolog of the Drosophila white gene is induced in monocyte-derived macrophages during cholesterol influx mediated by acetylated low-density lipoprotein. It is possible that human ABCG1 forms heterodimers with several heterologous partners.
Pssm-ID: 213201 [Multi-domain] Cd Length: 226 Bit Score: 110.44 E-value: 1.04e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 918 GLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPP---TSGTAYILGKDIRSEMSsiRQNLGVCPQHNVLFDMLTVEEHI 994
Cdd:cd03234 25 DVSLHVESGQVMAILGSSGSGKTTLLDAISGRVEGggtTSGQILFNGQPRKPDQF--QKCVAYVRQDDILLPGLTVRETL 102
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 995 WFYARLKgLSEKHVKAEMEQMALDVGLPPSKLK----SKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRG 1070
Cdd:cd03234 103 TYTAILR-LPRKSSDAIRKKRVEDVLLRDLALTriggNLVKGISGGERRRVSIAVQLLWDPKVLILDEPTSGLDSFTALN 181
|
170 180 190 200
....*....|....*....|....*....|....*....|..
gi 568925948 1071 IWELLLKY-RQGRTIILSTHHmDEADI--LGDRIAIISHGKL 1109
Cdd:cd03234 182 LVSTLSQLaRRNRIVILTIHQ-PRSDLfrLFDRILLLSSGEI 222
|
|
| ABC_cobalt_CbiO_domain2 |
cd03226 |
Second domain of the ATP-binding cassette component of cobalt transport system; Domain II of ... |
900-1109 |
2.13e-26 |
|
Second domain of the ATP-binding cassette component of cobalt transport system; Domain II of the ABC component of a cobalt transport family found in bacteria, archaea, and eukaryota. The transition metal cobalt is an essential component of many enzymes and must be transported into cells in appropriate amounts when needed. The CbiMNQO family ABC transport system is involved in cobalt transport in association with the cobalamin (vitamin B12) biosynthetic pathways. Most cobalt (Cbi) transport systems possess a separate CbiN component, the cobalt-binding periplasmic protein, and they are encoded by the conserved gene cluster cbiMNQO. Both the CbiM and CbiQ proteins are integral cytoplasmic membrane proteins, and the CbiO protein has the linker peptide and the Walker A and B motifs commonly found in the ATPase components of the ABC-type transport systems.
Pssm-ID: 213193 [Multi-domain] Cd Length: 205 Bit Score: 108.88 E-value: 2.13e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-----RSEMSSIRQN 974
Cdd:cd03226 1 RIENISFSYKKGTEI-LDDLSLDLYAGEIIALTGKNGAGKTTLAKILAGLIKESSGSILLNGKPIkakerRKSIGYVMQD 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 lgvcPQHNVLFDmlTVEEHIwfYARLKGLSEKHVKAE--MEQMALdvglppSKLKSKTSQ-LSGGMQRKLSVALAFVGGS 1051
Cdd:cd03226 80 ----VDYQLFTD--SVREEL--LLGLKELDAGNEQAEtvLKDLDL------YALKERHPLsLSGGQKQRLAIAAALLSGK 145
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03226 146 DLLIFDEPTSGLDYKNMERVGELIRElAAQGKAVIVITHDYEFLAKVCDRVLLLANGAI 204
|
|
| ABC_NrtD_SsuB_transporters |
cd03293 |
ATP-binding cassette domain of the nitrate and sulfonate transporters; NrtD and SsuB are the ... |
1914-2119 |
5.19e-26 |
|
ATP-binding cassette domain of the nitrate and sulfonate transporters; NrtD and SsuB are the ATP-binding subunits of the bacterial ABC-type nitrate and sulfonate transport systems, respectively. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213260 [Multi-domain] Cd Length: 220 Bit Score: 108.33 E-value: 5.19e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYR--RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknSILSN---IHEVHQ 1988
Cdd:cd03293 1 LEVRNVSKTYGggGGAVTALEDISLSVEEGEFVALVGPSGCGKSTLLRIIAGLERPTSG-------EVLVDgepVTGPGP 73
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:cd03293 74 DRGYVFQQDALLPWLTVLDNVALGLELQGVPKAEARERAEELLELVGLSGFENAYPHQLSGGMRQRVALARALAVDPDVL 153
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIV-KEGRSVVLTSHSMEECEALCTRMAIM 2119
Cdd:cd03293 154 LLDEPFSALDALTREQLQEELLDIWrETGKTVLLVTHDIDEAVFLADRVVVL 205
|
|
| AbcC |
COG1135 |
ABC-type methionine transport system, ATPase component [Amino acid transport and metabolism]; |
899-1109 |
6.10e-26 |
|
ABC-type methionine transport system, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 440750 [Multi-domain] Cd Length: 339 Bit Score: 111.32 E-value: 6.10e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:COG1135 2 IELENLSKTFPTKGGpvTALDDVSLTIEKGEIFGIIGYSGAGKSTLIRCINLLERPTSGSVLVDGVDLtalsERELRAAR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVCPQH-NvLFDMLTVEEHIWFYARLKGLSekhvKAEMEQMA---LD-VGLPpSKLKSKTSQLSGGMQRKLSVALAF 1047
Cdd:COG1135 82 RKIGMIFQHfN-LLSSRTVAENVALPLEIAGVP----KAEIRKRVaelLElVGLS-DKADAYPSQLSGGQKQRVGIARAL 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG1135 156 ANNPKVLLCDEATSALDPETTRSILDLLKDINRelGLTIVLITHEMDVVRRICDRVAVLENGRI 219
|
|
| GsiA |
COG1123 |
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain ... |
1913-2142 |
6.21e-26 |
|
ABC-type glutathione transport system ATPase component, contains duplicated ATPase domain [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 440740 [Multi-domain] Cd Length: 514 Bit Score: 114.62 E-value: 6.21e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIY---RRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL----SNIHE 1985
Cdd:COG1123 260 LLEVRNLSKRYpvrGKGGVRAVDDVSLTLRRGETLGLVGESGSGKSTLARLLLGLLRPTSGSILFDGKDLTklsrRSLRE 339
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYCPQ--FDAITELLTGREHVEFFALLRGV-PEKEVGKVGEWAIRKLGLvkyGEKYASNY----SGGNKRKLSTA 2058
Cdd:COG1123 340 LRRRVQMVFQdpYSSLNPRMTVGDIIAEPLRLHGLlSRAERRERVAELLERVGL---PPDLADRYphelSGGQRQRVAIA 416
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2059 MALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRF 2137
Cdd:COG1123 417 RALALEPKLLILDEPTSALDVSVQAQILNLLRDLQRElGLTYLFISHDLAVVRYIADRVAVMYDGRIVEDGPTEEVFANP 496
|
....*
gi 568925948 2138 GDGYT 2142
Cdd:COG1123 497 QHPYT 501
|
|
| ABC_MetN_methionine_transporter |
cd03258 |
ATP-binding cassette domain of methionine transporter; MetN (also known as YusC) is an ... |
1913-2133 |
8.85e-26 |
|
ATP-binding cassette domain of methionine transporter; MetN (also known as YusC) is an ABC-type transporter encoded by metN of the metNPQ operon in Bacillus subtilis that is involved in methionine transport. Other members of this system include the MetP permease and the MetQ substrate binding protein. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213225 [Multi-domain] Cd Length: 233 Bit Score: 108.05 E-value: 8.85e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIY--RRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDAFLNKNSI--LSN--IHE 1985
Cdd:cd03258 1 MIELKNVSKVFgdTGGKVTALKDVSLSVPKGEIFGIIGRSGAGKSTLIRCINGlERP-TSGSVLVDGTDLtlLSGkeLRK 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYCPQ-FDaiteLLTGR---EHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMAL 2061
Cdd:cd03258 80 ARRRIGMIFQhFN----LLSSRtvfENVALPLEIAGVPKAEIEERVLELLELVGLEDKADAYPAQLSGGQKQRVGIARAL 155
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2062 IGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:cd03258 156 ANNPKVLLCDEATSALDPETTQSILALLRDINRElGLTIVLITHEMEVVKRICDRVAVMEKGEVVEEGTVEEV 228
|
|
| ABC_Iron-Siderophores_B12_Hemin |
cd03214 |
ATP-binding component of iron-siderophores, vitamin B12 and hemin transporters and related ... |
1915-2128 |
9.20e-26 |
|
ATP-binding component of iron-siderophores, vitamin B12 and hemin transporters and related proteins; ABC transporters, involved in the uptake of siderophores, heme, and vitamin B12, are widely conserved in bacteria and archaea. Only very few species lack representatives of the siderophore family transporters. The E. coli BtuCD protein is an ABC transporter mediating vitamin B12 uptake. The two ATP-binding cassettes (BtuD) are in close contact with each other, as are the two membrane-spanning subunits (BtuC); this arrangement is distinct from that observed for the E. coli lipid flippase MsbA. The BtuC subunits provide 20 transmembrane helices grouped around a translocation pathway that is closed to the cytoplasm by a gate region, whereas the dimer arrangement of the BtuD subunits resembles the ATP-bound form of the Rad50 DNA repair enzyme. A prominent cytoplasmic loop of BtuC forms the contact region with the ATP-binding cassette and represent a conserved motif among the ABC transporters.
Pssm-ID: 213181 [Multi-domain] Cd Length: 180 Bit Score: 105.98 E-value: 9.20e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYC 1993
Cdd:cd03214 1 EVENLS--VGYGGRTVLDDLSLSIEAGEIVGILGPNGAGKSTLLKTLAGLLKPSSGEILLDGKDLASlSPKELARKIAYV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQfdaitelltgrehveffallrgvpekevgkvgewAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03214 79 PQ----------------------------------ALELLGLAHLADRPFNELSGGERQRVLLARALAQEPPILLLDEP 124
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2074 TTGMDPKARRFLwncaLSIVKE-----GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:cd03214 125 TSHLDIAHQIEL----LELLRRlarerGKTVVMVLHDLNLAARYADRVILLKDGRIVAQG 180
|
|
| ABCC_bacteriocin_exporters |
cd03245 |
ATP-binding cassette domain of bacteriocin exporters, subfamily C; Many non-lantibiotic ... |
908-1109 |
9.82e-26 |
|
ATP-binding cassette domain of bacteriocin exporters, subfamily C; Many non-lantibiotic bacteriocins of lactic acid bacteria are produced as precursors which have N-terminal leader peptides that share similarities in amino acid sequence and contain a conserved processing site of two glycine residues in positions -1 and -2. A dedicated ATP-binding cassette (ABC) transporter is responsible for the proteolytic cleavage of the leader peptides and subsequent translocation of the bacteriocins across the cytoplasmic membrane.
Pssm-ID: 213212 [Multi-domain] Cd Length: 220 Bit Score: 107.29 E-value: 9.82e-26
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLF- 985
Cdd:cd03245 12 YPNQEIPALDNVSLTIRAGEKVAIIGRVGSGKSTLLKLLAGLYKPTSGSVLLDGTDIRQlDPADLRRNIGYVPQDVTLFy 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 ----DMLTV------EEHIWFYARLKGLSE---KHVKAemeqMALDVGlppsklkSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:cd03245 92 gtlrDNITLgapladDERILRAAELAGVTDfvnKHPNG----LDLQIG-------ERGRGLSGGQRQAVALARALLNDPP 160
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKL 1109
Cdd:cd03245 161 ILLLDEPTSAMDMNSEERLKERLRQLLGDKTLIIITHRPSLLD-LVDRIIVMDSGRI 216
|
|
| LivF |
COG0410 |
ABC-type branched-chain amino acid transport system, ATPase component LivF [Amino acid ... |
900-1108 |
1.20e-25 |
|
ABC-type branched-chain amino acid transport system, ATPase component LivF [Amino acid transport and metabolism];
Pssm-ID: 440179 [Multi-domain] Cd Length: 236 Bit Score: 107.76 E-value: 1.20e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSeMSS---IRQNLG 976
Cdd:COG0410 5 EVENLHAGY--GGIHVLHGVSLEVEEGEIVALLGRNGAGKTTLLKAISGLLPPRSGSIRFDGEDITG-LPPhriARLGIG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEH--IWFYARlkgLSEKHVKAEMEQMaldVGLPP---SKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:COG0410 82 YVPEGRRIFPSLTVEENllLGAYAR---RDRAEVRADLERV---YELFPrlkERRRQRAGTLSGGEQQMLAIGRALMSRP 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELL--LKyRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:COG0410 156 KLLLLDEPSLGLAPLIVEEIFEIIrrLN-REGVTILLVEQNARFALEIADRAYVLERGR 213
|
|
| ABC_Carb_Monos_II |
cd03215 |
Second domain of the ATP-binding cassette component of monosaccharide transport system; This ... |
915-1109 |
1.95e-25 |
|
Second domain of the ATP-binding cassette component of monosaccharide transport system; This family represents domain II of the carbohydrate uptake proteins that transport only monosaccharides (Monos). The Carb_Monos family is involved in the uptake of monosaccharides, such as pentoses (such as xylose, arabinose, and ribose) and hexoses (such as xylose, arabinose, and ribose), that cannot be broken down to simple sugars by hydrolysis. In members of Carb_Monos family the single hydrophobic gene product forms a homodimer, while the ABC protein represents a fusion of two nucleotide-binding domains. However, it is assumed that two copies of the ABC domains are present in the assembled transporter.
Pssm-ID: 213182 [Multi-domain] Cd Length: 182 Bit Score: 105.21 E-value: 1.95e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC-----PQHNVLFDMLT 989
Cdd:cd03215 15 AVRDVSFEVRAGEIVGIAGLVGNGQTELAEALFGLRPPASGEITLDGKPVTRRSPRDAIRAGIAyvpedRKREGLVLDLS 94
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 990 VEEHIwfyarlkglsekhvkaemeqmALdvglppsklkskTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRR 1069
Cdd:cd03215 95 VAENI---------------------AL------------SSLLSGGNQQKVVLARWLARDPRVLILDEPTRGVDVGAKA 141
|
170 180 190 200
....*....|....*....|....*....|....*....|.
gi 568925948 1070 GIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03215 142 EIYRLIRELAdAGKAVLLISSELDELLGLCDRILVMYEGRI 182
|
|
| ABC_NikE_OppD_transporters |
cd03257 |
ATP-binding cassette domain of nickel/oligopeptides specific transporters; The ABC transporter ... |
1913-2123 |
3.43e-25 |
|
ATP-binding cassette domain of nickel/oligopeptides specific transporters; The ABC transporter subfamily specific for the transport of dipeptides, oligopeptides (OppD), and nickel (NikDE). The NikABCDE system of E. coli belongs to this family and is composed of the periplasmic binding protein NikA, two integral membrane components (NikB and NikC), and two ATPase (NikD and NikE). The NikABCDE transporter is synthesized under anaerobic conditions to meet the increased demand for nickel resulting from hydrogenase synthesis. The molecular mechanism of nickel uptake in many bacteria and most archaea is not known. Many other members of this ABC family are also involved in the uptake of dipeptides and oligopeptides. The oligopeptide transport system (Opp) is a five-component ABC transport composed of a membrane-anchored substrate binding proteins (SRP), OppA, two transmembrane proteins, OppB and OppC, and two ATP-binding domains, OppD and OppF.
Pssm-ID: 213224 [Multi-domain] Cd Length: 228 Bit Score: 106.05 E-value: 3.43e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEV---- 1986
Cdd:cd03257 1 LLEVKNLSVSFPTGGGSvkALDDVSFSIKKGETLGLVGESGSGKSTLARAILGLLKPTSGSIIFDGKDLLKLSRRLrkir 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNMGYCPQfDAITEL---LTGREHVE--FFALLRGVPEKEVGKVGEWAIRKLGLVK-YGEKYASNYSGGNKRKLSTAMA 2060
Cdd:cd03257 81 RKEIQMVFQ-DPMSSLnprMTIGEQIAepLRIHGKLSKKEARKEAVLLLLVGVGLPEeVLNRYPHELSGGQRQRVAIARA 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03257 160 LALNPKLLIADEPTSALDVSVQAQILDLLKKLQEElGLTLLFITHDLGVVAKIADRVAVMYAGK 223
|
|
| LivG |
COG0411 |
ABC-type branched-chain amino acid transport system, ATPase component LivG [Amino acid ... |
1913-2123 |
3.82e-25 |
|
ABC-type branched-chain amino acid transport system, ATPase component LivG [Amino acid transport and metabolism];
Pssm-ID: 440180 [Multi-domain] Cd Length: 257 Bit Score: 107.05 E-value: 3.82e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIY---RrkrkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEV-- 1986
Cdd:COG0411 4 LLEVRGLTKRFgglV-----AVDDVSLEVERGEIVGLIGPNGAGKTTLFNLITGFYRPTSGRILFDGRDITGlPPHRIar 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 ------HQNMGYCPQFDAITELLTG---REHVEFFALLRGVP-----EKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNK 2052
Cdd:COG0411 79 lgiartFQNPRLFPELTVLENVLVAahaRLGRGLLAALLRLPrarreEREARERAEELLERVGLADRADEPAGNLSYGQQ 158
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2053 RKLSTAMALIGGPPVVFLDEPTTGMDP--KAR--RFLwncaLSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG0411 159 RRLEIARALATEPKLLLLDEPAAGLNPeeTEElaELI----RRLRDErGITILLIEHDMDLVMGLADRIVVLDFGR 230
|
|
| ABCG_EPDR |
cd03213 |
Eye pigment and drug resistance transporter subfamily G of the ATP-binding cassette ... |
923-1109 |
5.00e-25 |
|
Eye pigment and drug resistance transporter subfamily G of the ATP-binding cassette superfamily; ABCG transporters are involved in eye pigment (EP) precursor transport, regulation of lipid-trafficking mechanisms, and pleiotropic drug resistance (DR). DR is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. Compared to other members of the ABC transporter subfamilies, the ABCG transporter family is composed of proteins that have an ATP-binding cassette domain at the N-terminus and a TM (transmembrane) domain at the C-terminus.
Pssm-ID: 213180 [Multi-domain] Cd Length: 194 Bit Score: 104.56 E-value: 5.00e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 923 FYEGQITSFLGHNGAGKTTTMSILTGL--FPPTSGTAYILGKDirSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWFYARL 1000
Cdd:cd03213 32 AKPGELTAIMGPSGAGKSTLLNALAGRrtGLGVSGEVLINGRP--LDKRSFRKIIGYVPQDDILHPTLTVRETLMFAAKL 109
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 KGLSekhvkaemeqmaldvglppsklksktsqlsGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ 1080
Cdd:cd03213 110 RGLS------------------------------GGERKRVSIALELVSNPSLLFLDEPTSGLDSSSALQVMSLLRRLAD 159
|
170 180 190
....*....|....*....|....*....|..
gi 568925948 1081 -GRTIILSTHHMdEADILG--DRIAIISHGKL 1109
Cdd:cd03213 160 tGRTIICSIHQP-SSEIFElfDKLLLLSQGRV 190
|
|
| cbiO |
PRK13636 |
cobalt transporter ATP-binding subunit; Provisional |
908-1130 |
8.30e-25 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184196 [Multi-domain] Cd Length: 283 Bit Score: 106.86 E-value: 8.30e-25
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI---RSEMSSIRQNLGVCPQH--N 982
Cdd:PRK13636 15 YSDGTH-ALKGININIKKGEVTAILGGNGAGKSTLFQNLNGILKPSSGRILFDGKPIdysRKGLMKLRESVGMVFQDpdN 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 983 VLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSkTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:PRK13636 94 QLFSA-SVYQDVSFGAVNLKLPEDEVRKRVDNALKRTGIEHLKDKP-THCLSFGQKKRVAIAGVLVMEPKVLVLDEPTAG 171
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1063 VDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVG--SSLFLKNQLGTGYYLTL 1130
Cdd:PRK13636 172 LDPMGVSEIMKLLVEMQKelGLTIIIATHDIDIVPLYCDNVFVMKEGRVILQGnpKEVFAEKEMLRKVNLRL 243
|
|
| ABC_Metallic_Cations |
cd03235 |
ATP-binding cassette domain of the metal-type transporters; This family includes transporters ... |
1915-2128 |
1.03e-24 |
|
ATP-binding cassette domain of the metal-type transporters; This family includes transporters involved in the uptake of various metallic cations such as iron, manganese, and zinc. The ATPases of this group of transporters are very similar to members of iron-siderophore uptake family suggesting that they share a common ancestor. The best characterized metal-type ABC transporters are the YfeABCD system of Y. pestis, the SitABCD system of Salmonella enterica serovar Typhimurium, and the SitABCD transporter of Shigella flexneri. Moreover other uncharacterized homologs of these metal-type transporters are mainly found in pathogens like Haemophilus or enteroinvasive E. coli isolates.
Pssm-ID: 213202 [Multi-domain] Cd Length: 213 Bit Score: 104.15 E-value: 1.03e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilsNIHEVHQNMGYCP 1994
Cdd:cd03235 1 EVEDLT--VSYGGHPVLEDVSFEVKPGEFLAIVGPNGAGKSTLLKAILGLLKPTSGSIRVFGK----PLEKERKRIGYVP 74
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1995 QFDAI--TELLTGREHVE-----FFALLRGVPEKEVGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:cd03235 75 QRRSIdrDFPISVRDVVLmglygHKGLFRRLSKADKAKVDE-ALERVGLSELADRQIGELSGGQQQRVLLARALVQDPDL 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2068 VFLDEPTTGMDPKARRFLwncaLSIVK----EGRSVVLTSHSMEECEALCTRmAIMVNGRFRCLG 2128
Cdd:cd03235 154 LLLDEPFAGVDPKTQEDI----YELLRelrrEGMTILVVTHDLGLVLEYFDR-VLLLNRTVVASG 213
|
|
| ABC2_perm_RbbA |
NF033858 |
ribosome-associated ATPase/putative transporter RbbA; |
1930-2123 |
1.38e-24 |
|
ribosome-associated ATPase/putative transporter RbbA;
Pssm-ID: 468210 [Multi-domain] Cd Length: 907 Bit Score: 112.53 E-value: 1.38e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQ-FDAITElLTGREH 2008
Cdd:NF033858 281 AVDHVSFRIRRGEIFGFLGSNGCGKSTTMKMLTGLLPASEGEAWLFGQPVDAGDIATRRRVGYMSQaFSLYGE-LTVRQN 359
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2009 VEFFALLRGVPEKEV-GKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWN 2087
Cdd:NF033858 360 LELHARLFHLPAAEIaARVAE-MLERFDLADVADALPDSLPLGIRQRLSLAVAVIHKPELLILDEPTSGVDPVARDMFWR 438
|
170 180 190
....*....|....*....|....*....|....*...
gi 568925948 2088 --CALSiVKEGRSVVLTSHSMEECEaLCTRMAIMVNGR 2123
Cdd:NF033858 439 llIELS-REDGVTIFISTHFMNEAE-RCDRISLMHAGR 474
|
|
| ABC_HisP_GlnQ |
cd03262 |
ATP-binding cassette domain of the histidine and glutamine transporters; HisP and GlnQ are the ... |
899-1109 |
2.60e-24 |
|
ATP-binding cassette domain of the histidine and glutamine transporters; HisP and GlnQ are the ATP-binding components of the bacterial periplasmic histidine and glutamine permeases, respectively. Histidine permease is a multi-subunit complex containing the HisQ and HisM integral membrane subunits and two copies of HisP. HisP has properties intermediate between those of integral and peripheral membrane proteins and is accessible from both sides of the membrane, presumably by its interaction with HisQ and HisM. The two HisP subunits form a homodimer within the complex. The domain structure of the amino acid uptake systems is typical for prokaryotic extracellular solute binding protein-dependent uptake systems. All of the amino acid uptake systems also have at least one, and in a few cases, two extracellular solute binding proteins located in the periplasm of Gram-negative bacteria, or attached to the cell membrane of Gram-positive bacteria. The best-studied member of the PAAT (polar amino acid transport) family is the HisJQMP system of S. typhimurium, where HisJ is the extracellular solute binding proteins and HisP is the ABC protein.
Pssm-ID: 213229 [Multi-domain] Cd Length: 213 Bit Score: 102.99 E-value: 2.60e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI---RSEMSSIRQNL 975
Cdd:cd03262 1 IEIKNLHKSF--GDFHVLKGIDLTVKKGEVVVIIGPSGSGKSTLLRCINLLEEPDSGTIIIDGLKLtddKKNINELRQKV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQHNVLFDMLTVEEHIWFYAR-LKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:cd03262 79 GMVFQQFNLFPHLTVLENITLAPIkVKGMSKAEAEERALELLEKVGLA-DKADAYPAQLSGGQQQRVAIARALAMNPKVM 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03262 158 LFDEPTSALDPELVGEVLDVMKDLaEEGMTMVVVTHEMGFAREVADRVIFMDDGRI 213
|
|
| PhnK |
COG1101 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
899-1108 |
2.66e-24 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 440718 [Multi-domain] Cd Length: 264 Bit Score: 104.78 E-value: 2.66e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGM---KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-------RSEM 968
Cdd:COG1101 2 LELKNLSKTFNPGTvneKRALDGLNLTIEEGDFVTVIGSNGAGKSTLLNAIAGSLPPDSGSILIDGKDVtklpeykRAKY 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 -SSIRQN--LGVCPQhnvlfdmLTVEEHIWF-YARLK------GLSEKHVKAEMEQMA-LDVGLPpSKLKSKTSQLSGGm 1037
Cdd:COG1101 82 iGRVFQDpmMGTAPS-------MTIEENLALaYRRGKrrglrrGLTKKRRELFRELLAtLGLGLE-NRLDTKVGLLSGG- 152
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1038 QRK-LSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:COG1101 153 QRQaLSLLMATLTKPKLLLLDEHTAALDPKTAALVLELTEKIveENNLTTLMVTHNMEQALDYGNRLIMMHEGR 226
|
|
| MglA |
COG1129 |
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism]; |
899-1114 |
3.16e-24 |
|
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440745 [Multi-domain] Cd Length: 497 Bit Score: 109.34 E-value: 3.16e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRseMSSIR--QNLG 976
Cdd:COG1129 5 LEMRGISKSF-GGVK-ALDGVSLELRPGEVHALLGENGAGKSTLMKILSGVYQPDSGEILLDGEPVR--FRSPRdaQAAG 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VC--PQHNVLFDMLTVEEHIWF---YARLKGLSEKHVKAEMEQM--ALDVGLPPSKlksKTSQLSGGMQRKLSVALAFVG 1049
Cdd:COG1129 81 IAiiHQELNLVPNLSVAENIFLgrePRRGGLIDWRAMRRRARELlaRLGLDIDPDT---PVGDLSVAQQQLVEIARALSR 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1050 GSKVVILDEPTAgvdPYSRRGIWELL-----LKyRQGRTIILSTHHMDEADILGDRIAIISHGKLccVGS 1114
Cdd:COG1129 158 DARVLILDEPTA---SLTEREVERLFriirrLK-AQGVAIIYISHRLDEVFEIADRVTVLRDGRL--VGT 221
|
|
| ModF |
COG1119 |
ABC-type molybdenum transport system, ATPase component ModF/photorepair protein PhrA ... |
899-1093 |
3.25e-24 |
|
ABC-type molybdenum transport system, ATPase component ModF/photorepair protein PhrA [Inorganic ion transport and metabolism];
Pssm-ID: 440736 [Multi-domain] Cd Length: 250 Bit Score: 104.01 E-value: 3.25e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNlVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSG-TAYILGKDIRSE-MSSIRQNLG 976
Cdd:COG1119 4 LELRN-VTVRRGG-KTILDDISWTVKPGEHWAILGPNGAGKSTLLSLITGDLPPTYGnDVRLFGERRGGEdVWELRKRIG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCpqHNVLFDMLTVEEHIW------FYARLkGLSeKHVKAEMEQMALD----VGLppSKLKSKT-SQLSGGMQRKLSVAL 1045
Cdd:COG1119 82 LV--SPALQLRFPRDETVLdvvlsgFFDSI-GLY-REPTDEQRERAREllelLGL--AHLADRPfGTLSQGEQRRVLIAR 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 1046 AFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDE 1093
Cdd:COG1119 156 ALVKDPELLILDEPTAGLDLGARELLLALLdkLAAEGAPTLVLVTHHVEE 205
|
|
| TauB |
COG1116 |
ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component [Inorganic ion ... |
1913-2119 |
4.71e-24 |
|
ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 440733 [Multi-domain] Cd Length: 260 Bit Score: 103.63 E-value: 4.71e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRK--RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlsniHEVHQNM 1990
Cdd:COG1116 7 ALELRGVSKRFPTGggGVTALDDVSLTVAAGEFVALVGPSGCGKSTLLRLIAGLEKPTSGEVLVDGKPV----TGPGPDR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAiteLL---TGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:COG1116 83 GVVFQEPA---LLpwlTVLDNVALGLELRGVPKAERRERARELLELVGLAGFEDAYPHQLSGGMRQRVAIARALANDPEV 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIM 2119
Cdd:COG1116 160 LLMDEPFGALDALTRERLQDELLRLWQEtGKTVLFVTHDVDEAVFLADRVVVL 212
|
|
| metN |
PRK11153 |
DL-methionine transporter ATP-binding subunit; Provisional |
901-1109 |
5.78e-24 |
|
DL-methionine transporter ATP-binding subunit; Provisional
Pssm-ID: 236863 [Multi-domain] Cd Length: 343 Bit Score: 105.65 E-value: 5.78e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYRDGMK--VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQN 974
Cdd:PRK11153 4 LKNISKVFPQGGRtiHALNNVSLHIPAGEIFGVIGASGAGKSTLIRCINLLERPTSGRVLVDGQDLtalsEKELRKARRQ 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHnvlFDML---TVEEHIWFYARLKGLSEKHVKAEMEQMaLD-VGLppSKLKSK-TSQLSGGMQRKLSVALAFVG 1049
Cdd:PRK11153 84 IGMIFQH---FNLLssrTVFDNVALPLELAGTPKAEIKARVTEL-LElVGL--SDKADRyPAQLSGGQKQRVAIARALAS 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1050 GSKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK11153 158 NPKVLLCDEATSALDPATTRSILELLKDInrELGLTIVLITHEMDVVKRICDRVAVIDAGRL 219
|
|
| CydD |
TIGR02857 |
thiol reductant ABC exporter, CydD subunit; The gene pair cydCD encodes an ABC-family ... |
899-1095 |
6.12e-24 |
|
thiol reductant ABC exporter, CydD subunit; The gene pair cydCD encodes an ABC-family transporter in which each gene contains an N-terminal membrane-spanning domain (pfam00664) and a C-terminal ATP-binding domain (pfam00005). In E. coli these genes were discovered as mutants which caused the terminal heme-copper oxidase complex cytochrome bd to fail to assemble. Recent work has shown that the transporter is involved in export of redox-active thiol compounds such as cysteine and glutathione. The linkage to assembly of the cytochrome bd complex is further supported by the conserved operon structure found outside the gammaproteobacteria (cydABCD) containing both the transporter and oxidase genes components. The genes used as the seed members for this model are all either found in the gammproteobacterial context or the CydABCD context. All members of this family scoring above trusted at the time of its creation were from genomes which encode a cytochrome bd complex. Unfortunately, the gene symbol nomenclature adopted based on this operon in B. subtilis assigns cydC to the third gene in the operon where this gene is actually homologous to the E. coli cydD gene. We have chosen to name all homologs in this family in accordance with the precedence of publication of the E. coli name, CydD
Pssm-ID: 274323 [Multi-domain] Cd Length: 529 Bit Score: 108.53 E-value: 6.12e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:TIGR02857 322 LEFSGVSVAYPGRRP-ALRPVSFTVPPGERVALVGPSGAGKSTLLNLLLGFVDPTEGSIAVNGVPLADaDADSWRDQIAW 400
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHIWFYARlkGLSEKHVKAEMEQMALD---VGLPP---SKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:TIGR02857 401 VPQHPFLFAG-TIAENIRLARP--DASDAEIREALERAGLDefvAALPQgldTPIGEGGAGLSGGQAQRLALARAFLRDA 477
|
170 180 190 200
....*....|....*....|....*....|....*....|....*..
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTH---HMDEAD 1095
Cdd:TIGR02857 478 PLLLLDEPTAHLDAETEAEVLEALRALAQGRTVLLVTHrlaLAALAD 524
|
|
| ABC_NatA_like |
cd03267 |
ATP-binding cassette domain of an uncharacterized transporter similar in sequence to NatA; ... |
1914-2123 |
6.49e-24 |
|
ATP-binding cassette domain of an uncharacterized transporter similar in sequence to NatA; NatA is the ATPase component of a bacterial ABC-type Na+ transport system called NatAB, which catalyzes ATP-dependent electrogenic Na+ extrusion without mechanically coupled to proton or K+ uptake. NatB possess six putative membrane spanning regions at its C-terminus. In B. subtilis, NatAB is inducible by agents such as ethanol and protonophores, which lower the proton-motive force across the membrane. The closest sequence similarity to NatA is exhibited by DrrA of the two-component daunorubicin- and doxorubicin-efflux system. Hence, the functional NatAB is presumably assembled with two copies of the single ATP-binding protein and the single integral membrane protein.
Pssm-ID: 213234 [Multi-domain] Cd Length: 236 Bit Score: 102.80 E-value: 6.49e-24
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRK-------------------PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDA-- 1972
Cdd:cd03267 1 IEVSNLSKSYRVYSKepgligslkslfkrkyrevEALKGISFTIEKGEIVGFIGPNGAGKTTTLKILSGLLQPTSGEVrv 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1973 -----FLNKNSILSNIHEVhqnmgycpqFDAITELLTGREHVEFFALLR---GVPEKEVGKVGEWAIRKLGLVKYGEKYA 2044
Cdd:cd03267 81 aglvpWKRRKKFLRRIGVV---------FGQKTQLWWDLPVIDSFYLLAaiyDLPPARFKKRLDELSELLDLEELLDTPV 151
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2045 SNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03267 152 RQLSLGQRMRAEIAAALLHEPEILFLDEPTIGLDVVAQENIRNFLKEYNRErGTTVLLTSHYMKDIEALARRVLVIDKGR 231
|
|
| ABC2_perm_RbbA |
NF033858 |
ribosome-associated ATPase/putative transporter RbbA; |
899-1125 |
1.59e-23 |
|
ribosome-associated ATPase/putative transporter RbbA;
Pssm-ID: 468210 [Multi-domain] Cd Length: 907 Bit Score: 109.06 E-value: 1.59e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDirseMSSIRQNLGVC 978
Cdd:NF033858 2 ARLEGVSHRY--GKTVALDDVSLDIPAGCMVGLIGPDGVGKSSLLSLIAGARKIQQGRVEVLGGD----MADARHRRAVC 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQ---------HNvLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGL------PPSKLksktsqlSGGMQRKLSV 1043
Cdd:NF033858 76 PRiaympqglgKN-LYPTLSVFENLDFFGRLFGQDAAERRRRIDELLRATGLapfadrPAGKL-------SGGMKQKLGL 147
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGR---TIILSTHHMDEADILgDRIAIISHGKLCCVGSSLFLKN 1120
Cdd:NF033858 148 CCALIHDPDLLILDEPTTGVDPLSRRQFWELIDRIRAERpgmSVLVATAYMEEAERF-DWLVAMDAGRVLATGTPAELLA 226
|
....*
gi 568925948 1121 QLGTG 1125
Cdd:NF033858 227 RTGAD 231
|
|
| ABC_Pro_Gly_Betaine |
cd03294 |
ATP-binding cassette domain of the osmoprotectant proline/glycine betaine uptake system; This ... |
1914-2123 |
1.75e-23 |
|
ATP-binding cassette domain of the osmoprotectant proline/glycine betaine uptake system; This family comprises the glycine betaine/L-proline ATP binding subunit in bacteria and its equivalents in archaea. This transport system belong to the larger ATP-Binding Cassette (ABC) transporter superfamily. The characteristic feature of these transporters is the obligatory coupling of ATP hydrolysis to substrate translocation. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213261 [Multi-domain] Cd Length: 269 Bit Score: 102.34 E-value: 1.75e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIG----------------------IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGD 1971
Cdd:cd03294 1 IKIKGLYKIFGKNPQKAFKLLAKGkskeeilkktgqtvgvndvsldVREGEIFVIMGLSGSGKSTLLRCINRLIEPTSGK 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1972 AFLNKNSILS------------NIHEVHQNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKY 2039
Cdd:cd03294 81 VLIDGQDIAAmsrkelrelrrkKISMVFQSFALLPH-------RTVLENVAFGLEVQGVPRAEREERAAEALELVGLEGW 153
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2040 GEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAI 2118
Cdd:cd03294 154 EHKYPDELSGGMQQRVGLARALAVDPDILLMDEAFSALDPLIRREMQDELLRLQAElQKTIVFITHDLDEALRLGDRIAI 233
|
....*
gi 568925948 2119 MVNGR 2123
Cdd:cd03294 234 MKDGR 238
|
|
| DppD |
COG0444 |
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid ... |
899-1103 |
2.03e-23 |
|
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid transport and metabolism, Inorganic ion transport and metabolism];
Pssm-ID: 440213 [Multi-domain] Cd Length: 320 Bit Score: 103.59 E-value: 2.03e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVY--RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPP---TSGTAYILGKDI----RSEMS 969
Cdd:COG0444 2 LEVRNLKVYFptRRGVVKAVDGVSFDVRRGETLGLVGESGSGKSTLARAILGLLPPpgiTSGEILFDGEDLlklsEKELR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 970 SIRQN-LGVCPQhnvlfD-------MLTVEEHIW-FYARLKGLSEKHVKAEMEQMALDVGLPPSK--LKSKTSQLSGGMQ 1038
Cdd:COG0444 82 KIRGReIQMIFQ-----DpmtslnpVMTVGDQIAePLRIHGGLSKAEARERAIELLERVGLPDPErrLDRYPHELSGGMR 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1039 RKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAI 1103
Cdd:COG0444 157 QRVMIARALALEPKLLIADEPTTALDVTIQAQILNLLKDLQRelGLAILFITHDLGVVAEIADRVAV 223
|
|
| potG |
PRK11607 |
putrescine ABC transporter ATP-binding subunit PotG; |
901-1114 |
2.75e-23 |
|
putrescine ABC transporter ATP-binding subunit PotG;
Pssm-ID: 183226 [Multi-domain] Cd Length: 377 Bit Score: 104.53 E-value: 2.75e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYrDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVCPQ 980
Cdd:PRK11607 22 IRNLTKSF-DG-QHAVDDVSLTIYKGEIFALLGASGCGKSTLLRMLAGFEQPTAGQIMLDGVDL-SHVPPYQRPINMMFQ 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPT 1060
Cdd:PRK11607 99 SYALFPHMTVEQNIAFGLKQDKLPKAEIASRVNEMLGLVHMQEFA-KRKPHQLSGGQRQRVALARSLAKRPKLLLLDEPM 177
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1061 AGVDPYSR-RGIWELL-LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK11607 178 GALDKKLRdRMQLEVVdILERVGVTCVMVTHDQEEAMTMAGRIAIMNRGKFVQIGE 233
|
|
| DppF |
COG1124 |
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid ... |
1914-2142 |
2.82e-23 |
|
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid transport and metabolism, Inorganic ion transport and metabolism];
Pssm-ID: 440741 [Multi-domain] Cd Length: 248 Bit Score: 101.03 E-value: 2.82e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYR--RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHEVHQNM 1990
Cdd:COG1124 2 LEVRNLSVSYGqgGRRVPVLKDVSLEVAPGESFGLVGESGSGKSTLLRALAGLERPWSGEVTFDGRPVtRRRRKAFRRRV 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQfDAITEL---LTGREHVEFFALLRGVPEKEVgKVGEwAIRKLGLvkyGEKYASNY----SGGNKRKLSTAMALIG 2063
Cdd:COG1124 82 QMVFQ-DPYASLhprHTVDRILAEPLRIHGLPDREE-RIAE-LLEQVGL---PPSFLDRYphqlSGGQRQRVAIARALIL 155
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2064 GPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGYT 2142
Cdd:COG1124 156 EPELLLLDEPTSALDVSVQAEILNLLKDLREErGLTYLFVSHDLAVVAHLCDRVAVMQNGRIVEELTVADLLAGPKHPYT 235
|
|
| ABC_ThiQ_thiamine_transporter |
cd03298 |
ATP-binding cassette domain of the thiamine transport system; Part of the ... |
919-1113 |
2.91e-23 |
|
ATP-binding cassette domain of the thiamine transport system; Part of the binding-protein-dependent transport system tbpA-thiPQ for thiamine and TPP. Probably responsible for the translocation of thiamine across the membrane. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213265 [Multi-domain] Cd Length: 211 Bit Score: 99.87 E-value: 2.91e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 919 LALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWF-- 996
Cdd:cd03298 17 FDLTFAQGEITAIVGPSGSGKSTLLNLIAGFETPQSGRVLINGVDV-TAAPPADRPVSMLFQENNLFAHLTVEQNVGLgl 95
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 997 --YARLKGLSEKHVKAEMEQMALDvglppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWEL 1074
Cdd:cd03298 96 spGLKLTAEDRQAIEVALARVGLA-----GLEKRLPGELSGGERQRVALARVLVRDKPVLLLDEPFAALDPALRAEMLDL 170
|
170 180 190 200
....*....|....*....|....*....|....*....|.
gi 568925948 1075 LLKYRQGR--TIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:cd03298 171 VLDLHAETkmTVLMVTHQPEDAKRLAQRVVFLDNGRIAAQG 211
|
|
| LptB |
COG1137 |
ABC-type lipopolysaccharide export system, ATPase component [Cell wall/membrane/envelope ... |
1913-2123 |
3.68e-23 |
|
ABC-type lipopolysaccharide export system, ATPase component [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440752 [Multi-domain] Cd Length: 240 Bit Score: 100.49 E-value: 3.68e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSN--IHE-VHQN 1989
Cdd:COG1137 3 TLEAENLVKSY--GKRTVVKDVSLEVNQGEIVGLLGPNGAGKTTTFYMIVGLVKPDSGRIFLDGEDI-THlpMHKrARLG 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:COG1137 80 IGYLPQEASIFRKLTVEDNILAVLELRKLSKKEREERLEELLEEFGITHLRKSKAYSLSGGERRRVEIARALATNPKFIL 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2070 LDEPTTGMDPKA----RRflwncalsIVKE----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG1137 160 LDEPFAGVDPIAvadiQK--------IIRHlkerGIGVLITDHNVRETLGICDRAYIISEGK 213
|
|
| ABC_MalK_N |
cd03301 |
The N-terminal ATPase domain of the maltose transporter, MalK; ATP binding cassette (ABC) ... |
1914-2128 |
4.37e-23 |
|
The N-terminal ATPase domain of the maltose transporter, MalK; ATP binding cassette (ABC) proteins function from bacteria to human, mediating the translocation of substances into and out of cells or organelles. ABC transporters contain two transmembrane-spanning domains (TMDs) or subunits and two nucleotide binding domains (NBDs) or subunits that couple transport to the hydrolysis of ATP. In the maltose transport system, the periplasmic maltose binding protein (MBP) stimulates the ATPase activity of the membrane-associated transporter, which consists of two transmembrane subunits, MalF and MalG, and two copies of the ATP binding subunit, MalK, and becomes tightly bound to the transporter in the catalytic transition state, ensuring that maltose is passed to the transporter as ATP is hydrolyzed.
Pssm-ID: 213268 [Multi-domain] Cd Length: 213 Bit Score: 99.64 E-value: 4.37e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS------NIHEVH 1987
Cdd:cd03301 1 VELENVTKRF--GNVTALDDLNLDIADGEFVVLLGPSGCGKTTTLRMIAGLEEPTSGRIYIGGRDVTDlppkdrDIAMVF 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:cd03301 79 QNYALYPH-------MTVYDNIAFGLKLRKVPKDEIDERVREVAELLQIEHLLDRKPKQLSGGQRQRVALGRAIVREPKV 151
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:cd03301 152 FLMDEPLSNLDAKLRVQMRAELKRLQQRlGTTTIYVTHDQVEAMTMADRIAVMNDGQIQQIG 213
|
|
| FetA |
COG4619 |
ABC-type iron transporter FetAB, ATPase component [Inorganic ion transport and metabolism]; |
1914-2124 |
4.72e-23 |
|
ABC-type iron transporter FetAB, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443661 [Multi-domain] Cd Length: 209 Bit Score: 99.51 E-value: 4.72e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:COG4619 1 LELEGLS--FRVGGKPILSPVSLTLEAGECVAITGPSGSGKSTLLRALADLDPPTSGEIYLDGKPLSAmPPPEWRRQVAY 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQ----FDAitellTGREHVEFFALLRGVPEKEvGKVGEWaIRKLGL-VKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:COG4619 79 VPQepalWGG-----TVRDNLPFPFQLRERKFDR-ERALEL-LERLGLpPDILDKPVERLSGGERQRLALIRALLLQPDV 151
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:COG4619 152 LLLDEPTSALDPENTRRVEELLREYLAEeGRAVLWVSHDPEQIERVADRVLTLEAGRL 209
|
|
| ABC_PotA_N |
cd03300 |
ATP-binding cassette domain of the polyamine transporter; PotA is an ABC-type transporter and ... |
1914-2137 |
4.92e-23 |
|
ATP-binding cassette domain of the polyamine transporter; PotA is an ABC-type transporter and the ATPase component of the spermidine/putrescine-preferential uptake system consisting of PotA, -B, -C, and -D. PotA has two domains with the N-terminal domain containing the ATPase activity and the residues required for homodimerization with PotA and heterdimerization with PotB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213267 [Multi-domain] Cd Length: 232 Bit Score: 100.00 E-value: 4.92e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS------NIHEVH 1987
Cdd:cd03300 1 IELENVSKFY--GGFVALDGVSLDIKEGEFFTLLGPSGCGKTTLLRLIAGFETPTSGEILLDGKDITNlpphkrPVNTVF 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:cd03300 79 QNYALFPH-------LTVFENIAFGLRLKKLPKAEIKERVAEALDLVQLEGYANRKPSQLSGGQQQRVAIARALVNEPKV 151
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL----KNRF 2137
Cdd:cd03300 152 LLLDEPLGALDLKLRKDMQLELKRLQKElGITFVFVTHDQEEALTMSDRIAVMNKGKIQQIGTPEEIyeepANRF 226
|
|
| 40850658_otr |
NF000106 |
oxytetracycline efflux ABC transporter Otr(C) ATP-binding subunit; |
1902-2148 |
5.49e-23 |
|
oxytetracycline efflux ABC transporter Otr(C) ATP-binding subunit;
Pssm-ID: 411078 [Multi-domain] Cd Length: 351 Bit Score: 102.89 E-value: 5.49e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1902 QRILDGGGQNDIlEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAG--KSTTFKMLTGDTPVTRGDAFLnknSI 1979
Cdd:NF000106 3 RKTISNGARNAV-EVRGLVKHFGEVK--AVDGVDLDVREGTVLGVLGP*GAA**RGALPAHV*GPDAGRRPWRF*---TW 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1980 LSNIHEVHQNMG-YCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTA 2058
Cdd:NF000106 77 CANRRALRRTIG*HRPVR*GRRESFSGRENLYMIGR*LDLSRKDARARADELLERFSLTEAAGRAAAKYSGGMRRRLDLA 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2059 MALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFG 2138
Cdd:NF000106 157 ASMIGRPAVLYLDEPTTGLDPRTRNEVWDEVRSMVRDGATVLLTTQYMEEAEQLAHELTVIDRGRVIADGKVDELKTKVG 236
|
250
....*....|
gi 568925948 2139 dGYTIVVRIA 2148
Cdd:NF000106 237 -GRTLQIRPA 245
|
|
| hmuV |
PRK13548 |
hemin importer ATP-binding subunit; Provisional |
900-1114 |
6.40e-23 |
|
hemin importer ATP-binding subunit; Provisional
Pssm-ID: 237422 [Multi-domain] Cd Length: 258 Bit Score: 100.62 E-value: 6.40e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVkvYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS----EMSSIRqnl 975
Cdd:PRK13548 4 EARNLS--VRLGGRTLLDDVSLTLRPGEVVAILGPNGAGKSTLLRALSGELSPDSGEVRLNGRPLADwspaELARRR--- 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQHNVL-FDmLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLppSKLKSKT-SQLSGG-MQRklsVALAFV---- 1048
Cdd:PRK13548 79 AVLPQHSSLsFP-FTVEEVVAMGRAPHGLSRAEDDALVAAALAQVDL--AHLAGRDyPQLSGGeQQR---VQLARVlaql 152
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1049 ----GGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13548 153 wepdGPPRWLLLDEPTSALDLAHQHHVLRLArqLAHERGLAVIVVLHDLNLAARYADRIVLLHQGRLVADGT 224
|
|
| ntrCD |
TIGR01184 |
nitrate transport ATP-binding subunits C and D; This model describes the ATP binding subunits ... |
916-1116 |
8.59e-23 |
|
nitrate transport ATP-binding subunits C and D; This model describes the ATP binding subunits of nitrate transport in bacteria and archaea. This protein belongs to the ATP-binding cassette (ABC) superfamily. It is thought that the two subunits encoded by ntrC and ntrD form the binding surface for interaction with ATP. This model is restricted in identifying ATP binding subunit associated with the nitrate transport. Nitrate assimilation is aided by other proteins derived from the operon which among others include products of ntrA - a regulatory protein; ntrB - a hydropbobic transmembrane permease and narB - a reductase. [Transport and binding proteins, Anions, Transport and binding proteins, Other]
Pssm-ID: 130252 [Multi-domain] Cd Length: 230 Bit Score: 99.46 E-value: 8.59e-23
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQnlgVCPQHNVLFDMLTVEEHIW 995
Cdd:TIGR01184 1 LKGVNLTIQQGEFISLIGHSGCGKSTLLNLISGLAQPTSGGVILEGKQI-TEPGPDRM---VVFQNYSLLPWLTVRENIA 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 996 FYAR--LKGLSEKHVKAEMEQMALDVGLPPSKLKsKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWE 1073
Cdd:TIGR01184 77 LAVDrvLPDLSKSERRAIVEEHIALVGLTEAADK-RPGQLSGGMKQRVAIARALSIRPKVLLLDEPFGALDALTRGNLQE 155
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 1074 LLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSL 1116
Cdd:TIGR01184 156 ELMQIWEehRVTVLMVTHDVDEALLLSDRVVMLTNGPAANIGQIL 200
|
|
| ABCC_MRP_domain2 |
cd03244 |
ATP-binding cassette domain 2 of multidrug resistance-associated protein; The ABC subfamily C ... |
899-1110 |
1.85e-22 |
|
ATP-binding cassette domain 2 of multidrug resistance-associated protein; The ABC subfamily C is also known as MRP (multidrug resistance-associated protein). Some of the MRP members have five additional transmembrane segments in their N-terminus, but the function of these additional membrane-spanning domains is not clear. The MRP was found in the multidrug-resistance lung cancer cell in which p-glycoprotein was not overexpressed. MRP exports glutathione by drug stimulation, as well as, certain substrates in conjugated forms with anions, such as glutathione, glucuronate, and sulfate.
Pssm-ID: 213211 [Multi-domain] Cd Length: 221 Bit Score: 97.95 E-value: 1.85e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:cd03244 3 IEFKNVSLRYRPNLPPVLKNISFSIKPGEKVGIVGRTGSGKSSLLLALFRLVELSSGSILIDGVDISKiGLHDLRSRISI 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTV-----------EEHIWfyarlKGLSEKHVKAEMEQMA--LDvglppSKLKSKTSQLSGGmQRKL-SV 1043
Cdd:cd03244 83 IPQDPVLFSG-TIrsnldpfgeysDEELW-----QALERVGLKEFVESLPggLD-----TVVEEGGENLSVG-QRQLlCL 150
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEadILG-DRIAIISHGKLC 1110
Cdd:cd03244 151 ARALLRKSKILVLDEATASVDPETDALIQKTIREAFKDCTVLTIAHRLDT--IIDsDRILVLDKGRVV 216
|
|
| ABC_OpuCA_Osmoprotection |
cd03295 |
ATP-binding cassette domain of the osmoprotectant transporter; OpuCA is a the ATP binding ... |
1914-2133 |
2.59e-22 |
|
ATP-binding cassette domain of the osmoprotectant transporter; OpuCA is a the ATP binding component of a bacterial solute transporter that serves a protective role to cells growing in a hyperosmolar environment. ABC (ATP-binding cassette) transporter nucleotide-binding domain; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition, to the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213262 [Multi-domain] Cd Length: 242 Bit Score: 98.14 E-value: 2.59e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:cd03295 1 IEFENVTKRYG-GGKKAVNNLNLEIAKGEFLVLIGPSGSGKTTTMKMINRLIEPTSGEIFIDGEDIREqDPVELRRKIGY 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGL--VKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:cd03295 80 VIQQIGLFPHMTVEENIALVPKLLKWPKEKIRERADELLALVGLdpAEFADRYPHELSGGQQQRVGVARALAADPPLLLM 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:cd03295 160 DEPFGALDPITRDQLQEEFKRLQQElGKTIVFVTHDIDEAFRLADRIAIMKNGEIVQVGTPDEI 223
|
|
| ABC_cobalt_CbiO_domain2 |
cd03226 |
Second domain of the ATP-binding cassette component of cobalt transport system; Domain II of ... |
1915-2122 |
2.62e-22 |
|
Second domain of the ATP-binding cassette component of cobalt transport system; Domain II of the ABC component of a cobalt transport family found in bacteria, archaea, and eukaryota. The transition metal cobalt is an essential component of many enzymes and must be transported into cells in appropriate amounts when needed. The CbiMNQO family ABC transport system is involved in cobalt transport in association with the cobalamin (vitamin B12) biosynthetic pathways. Most cobalt (Cbi) transport systems possess a separate CbiN component, the cobalt-binding periplasmic protein, and they are encoded by the conserved gene cluster cbiMNQO. Both the CbiM and CbiQ proteins are integral cytoplasmic membrane proteins, and the CbiO protein has the linker peptide and the Walker A and B motifs commonly found in the ATPase components of the ABC-type transport systems.
Pssm-ID: 213193 [Multi-domain] Cd Length: 205 Bit Score: 96.94 E-value: 2.62e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-----LSNIHEVHQN 1989
Cdd:cd03226 1 RIENISFSYKKGTE-ILDDLSLDLYAGEIIALTGKNGAGKTTLAKILAGLIKESSGSILLNGKPIkakerRKSIGYVMQD 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYcpQFdaitelltGREHVeFFALLRGVPEK-EVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:cd03226 80 VDY--QL--------FTDSV-REELLLGLKELdAGNEQAETVLKDLDLYALKERHPLSLSGGQKQRLAIAAALLSGKDLL 148
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNG 2122
Cdd:cd03226 149 IFDEPTSGLDYKNMERVGELIRELAAQGKAVIVITHDYEFLAKVCDRVLLLANG 202
|
|
| ABC_MTABC3_MDL1_MDL2 |
cd03249 |
ATP-binding cassette domain of a mitochondrial protein MTABC3 and related proteins; MTABC3 ... |
909-1133 |
2.69e-22 |
|
ATP-binding cassette domain of a mitochondrial protein MTABC3 and related proteins; MTABC3 (also known as ABCB6) is a mitochondrial ATP-binding cassette protein involved in iron homeostasis and one of four ABC transporters expressed in the mitochondrial inner membrane, the other three being MDL1(ABC7), MDL2, and ATM1. In fact, the yeast MDL1 (multidrug resistance-like protein 1) and MDL2 (multidrug resistance-like protein 2) transporters are also included in this CD. MDL1 is an ATP-dependent permease that acts as a high-copy suppressor of ATM1 and is thought to have a role in resistance to oxidative stress. Interestingly, subfamily B is more closely related to the carboxyl-terminal component of subfamily C than the two halves of ABCC molecules are with one another.
Pssm-ID: 213216 [Multi-domain] Cd Length: 238 Bit Score: 98.00 E-value: 2.69e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDM 987
Cdd:cd03249 13 RPDVPI-LKGLSLTIPPGKTVALVGSSGCGKSTVVSLLERFYDPTSGEILLDGVDIRDlNLRWLRSQIGLVSQEPVLFDG 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 988 lTVEEHIwFYARLKGLSEKHVKAEMEQMALD--VGLP---PSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:cd03249 92 -TIAENI-RYGKPDATDEEVEEAAKKANIHDfiMSLPdgyDTLVGERGSQLSGGQKQRIAIARALLRNPKILLLDEATSA 169
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1063 VDPYSRRGIWELLLKYRQGRTIILSTHHMdeADILG-DRIAIISHGKLCCVGSSLFLKNQlgTGYYLTLVKK 1133
Cdd:cd03249 170 LDAESEKLVQEALDRAMKGRTTIVIAHRL--STIRNaDLIAVLQNGQVVEQGTHDELMAQ--KGVYAKLVKA 237
|
|
| ABC_PhnC_transporter |
cd03256 |
ATP-binding cassette domain of the binding protein-dependent phosphonate transport system; ... |
1914-2123 |
2.80e-22 |
|
ATP-binding cassette domain of the binding protein-dependent phosphonate transport system; Phosphonates are a class of organophosphorus compounds characterized by a chemically stable carbon-to-phosphorus (C-P) bond. Phosphonates are widespread among naturally occurring compounds in all kingdoms of wildlife, but only prokaryotic microorganisms are able to cleave this bond. Certain bacteria such as E. coli can use alkylphosphonates as a phosphorus source. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213223 [Multi-domain] Cd Length: 241 Bit Score: 98.02 E-value: 2.80e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHE-------- 1985
Cdd:cd03256 1 IEVENLSKTYPNGKK-ALKDVSLSINPGEFVALIGPSGAGKSTLLRCLNGLVEPTSGSVLIDGTDINKLKGKalrqlrrq 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 ---VHQNMGYCPQFDAITELLTGR--EHVEFFALLRGVPEKEVGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:cd03256 80 igmIFQQFNLIERLSVLENVLSGRlgRRSTWRSLFGLFPKEEKQRALA-ALERVGLLDKAYQRADQLSGGQQQRVAIARA 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03256 159 LMQQPKLILADEPVASLDPASSRQVMDLLKRINREeGITVIVSLHQVDLAREYADRIVGLKDGR 222
|
|
| PRK13633 |
PRK13633 |
energy-coupling factor transporter ATPase; |
902-1109 |
3.20e-22 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237453 [Multi-domain] Cd Length: 280 Bit Score: 99.01 E-value: 3.20e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 902 QNLVKVYRDG----MKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMS--SIRQNL 975
Cdd:PRK13633 8 KNVSYKYESNeestEKLALDDVNLEVKKGEFLVILGRNGSGKSTIAKHMNALLIPSEGKVYVDGLDTSDEENlwDIRNKA 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQH--NVLFDMLtVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK13633 88 GMVFQNpdNQIVATI-VEEDVAFGPENLGIPPEEIRERVDESLKKVGMYEYR-RHAPHLLSGGQKQRVAIAGILAMRPEC 165
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1054 VILDEPTAGVDPYSRR----GIWELLLKYrqGRTIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:PRK13633 166 IIFDEPTAMLDPSGRRevvnTIKELNKKY--GITIILITHYMEEA-VEADRIIVMDSGKV 222
|
|
| PRK14239 |
PRK14239 |
phosphate transporter ATP-binding protein; Provisional |
911-1115 |
3.70e-22 |
|
phosphate transporter ATP-binding protein; Provisional
Pssm-ID: 184585 [Multi-domain] Cd Length: 252 Bit Score: 97.92 E-value: 3.70e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKVAVDGLALNFYEGQITSFLGHNGAGKTT---TMSILTGLFPPTSGTAYIL--GKDI---RSEMSSIRQNLGVCPQHN 982
Cdd:PRK14239 16 NKKKALNSVSLDFYPNEITALIGPSGSGKSTllrSINRMNDLNPEVTITGSIVynGHNIyspRTDTVDLRKEIGMVFQQP 95
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 983 VLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMAL-------DVglpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:PRK14239 96 NPFPM-SIYENVVYGLRLKGIKDKQVLDEAVEKSLkgasiwdEV---KDRLHDSALGLSGGQQQRVCIARVLATSPKIIL 171
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1056 LDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS 1115
Cdd:PRK14239 172 LDEPTSALDPISAGKIEETLLGLKDDYTMLLVTRSMQQASRISDRTGFFLDGDLIEYNDT 231
|
|
| PRK13651 |
PRK13651 |
cobalt transporter ATP-binding subunit; Provisional |
899-1109 |
3.99e-22 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184210 [Multi-domain] Cd Length: 305 Bit Score: 99.39 E-value: 3.99e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKV---AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS----- 970
Cdd:PRK13651 3 IKVKNIVKIFNKKLPTelkALDNVSVEINQGEFIAIIGQTGSGKTTFIEHLNALLLPDTGTIEWIFKDEKNKKKTkekek 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 --------------------IRQNLGVCPQ--HNVLFDMlTVEEHIWFYARLKGLSekhvKAEMEQMALD----VGLPPS 1024
Cdd:PRK13651 83 vleklviqktrfkkikkikeIRRRVGVVFQfaEYQLFEQ-TIEKDIIFGPVSMGVS----KEEAKKRAAKyielVGLDES 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1025 KLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADILGDRIAI 1103
Cdd:PRK13651 158 YLQRSPFELSGGQKRRVALAGILAMEPDFLVFDEPTAGLDPQGVKEILEIFDNlNKQGKTIILVTHDLDNVLEWTKRTIF 237
|
....*.
gi 568925948 1104 ISHGKL 1109
Cdd:PRK13651 238 FKDGKI 243
|
|
| ABCC_Glucan_exporter_like |
cd03254 |
ATP-binding cassette domain of glucan transporter and related proteins, subfamily C; Glucan ... |
910-1107 |
4.66e-22 |
|
ATP-binding cassette domain of glucan transporter and related proteins, subfamily C; Glucan exporter ATP-binding protein. In A. tumefaciens cyclic beta-1, 2-glucan must be transported into the periplasmic space to exert its action as a virulence factor. This subfamily belongs to the MRP-like family and is involved in drug, peptide, and lipid export. The MRP-like family, similar to all ABC proteins, have a common four-domain core structure constituted by two membrane-spanning domains each composed of six transmembrane (TM) helices and two nucleotide-binding domains (NBD). ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213221 [Multi-domain] Cd Length: 229 Bit Score: 96.91 E-value: 4.66e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 910 DGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMl 988
Cdd:cd03254 13 DEKKPVLKDINFSIKPGETVAIVGPTGAGKTTLINLLMRFYDPQKGQILIDGIDIRDiSRKSLRSMIGVVLQDTFLFSG- 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIwFYARLKGLSEKHVKAEMEQMALDVG--LPP---SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGV 1063
Cdd:cd03254 92 TIMENI-RLGRPNATDEEVIEAAKEAGAHDFImkLPNgydTVLGENGGNLSQGERQLLAIARAMLRDPKILILDEATSNI 170
|
170 180 190 200
....*....|....*....|....*....|....*....|....*....
gi 568925948 1064 DPYSRRGIWELLLKYRQGRTIILSTHHMD---EAD--ILGDRIAIISHG 1107
Cdd:cd03254 171 DTETEKLIQEALEKLMKGRTSIIIAHRLStikNADkiLVLDDGKIIEEG 219
|
|
| ABCG_EPDR |
cd03213 |
Eye pigment and drug resistance transporter subfamily G of the ATP-binding cassette ... |
1917-2123 |
4.82e-22 |
|
Eye pigment and drug resistance transporter subfamily G of the ATP-binding cassette superfamily; ABCG transporters are involved in eye pigment (EP) precursor transport, regulation of lipid-trafficking mechanisms, and pleiotropic drug resistance (DR). DR is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. Compared to other members of the ABC transporter subfamilies, the ABCG transporter family is composed of proteins that have an ATP-binding cassette domain at the N-terminus and a TM (transmembrane) domain at the C-terminus.
Pssm-ID: 213180 [Multi-domain] Cd Length: 194 Bit Score: 96.08 E-value: 4.82e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1917 KELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG--DTPVTRGDAFLN-KNSILSNIHEVhqnMGYC 1993
Cdd:cd03213 11 VTVKSSPSKSGKQLLKNVSGKAKPGELTAIMGPSGAGKSTLLNALAGrrTGLGVSGEVLINgRPLDKRSFRKI---IGYV 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVpekevgkvgewairklglvkygekyasnySGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03213 88 PQDDILHPTLTVRETLMFAAKLRGL-----------------------------SGGERKRVSIALELVSNPSLLFLDEP 138
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHS-MEECEALCTRMAIMVNGR 2123
Cdd:cd03213 139 TSGLDSSSALQVMSLLRRLADTGRTIICSIHQpSSEIFELFDKLLLLSQGR 189
|
|
| met_CoM_red_A2 |
TIGR03269 |
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in ... |
1895-2124 |
6.63e-22 |
|
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in methanogenesis, methyl coenzyme M reductase, contains alpha, beta, and gamma chains. In older literature, the complex of alpha, beta, and gamma chains was termed component C, while this single chain protein was termed methyl coenzyme M reductase system component A2. [Energy metabolism, Methanogenesis]
Pssm-ID: 132313 [Multi-domain] Cd Length: 520 Bit Score: 102.19 E-value: 6.63e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1895 EDVRRERQRILdgggQNDILEIKELTKIY---RRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGD 1971
Cdd:TIGR03269 265 SEVEKECEVEV----GEPIIKVRNVSKRYisvDRGVVKAVDNVSLEVKEGEIFGIVGTSGAGKTTLSKIIAGVLEPTSGE 340
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1972 ---------------AFLNKNSILSNIHEVHQNMGYCPQFDaITELLTGREHVEF---FALLRGVPE-KEVGKVGEWAIR 2032
Cdd:TIGR03269 341 vnvrvgdewvdmtkpGPDGRGRAKRYIGILHQEYDLYPHRT-VLDNLTEAIGLELpdeLARMKAVITlKMVGFDEEKAEE 419
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2033 KLglvkygEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEA 2111
Cdd:TIGR03269 420 IL------DKYPDELSEGERHRVALAQVLIKEPRIVILDEPTGTMDPITKVDVTHSILKAREEmEQTFIIVSHDMDFVLD 493
|
250
....*....|...
gi 568925948 2112 LCTRMAIMVNGRF 2124
Cdd:TIGR03269 494 VCDRAALMRDGKI 506
|
|
| MdlB |
COG1132 |
ABC-type multidrug transport system, ATPase and permease component [Defense mechanisms]; |
908-1114 |
9.19e-22 |
|
ABC-type multidrug transport system, ATPase and permease component [Defense mechanisms];
Pssm-ID: 440747 [Multi-domain] Cd Length: 579 Bit Score: 102.17 E-value: 9.19e-22
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFD 986
Cdd:COG1132 349 YPGD-RPVLKDISLTIPPGETVALVGPSGSGKSTLVNLLLRFYDPTSGRILIDGVDIRDlTLESLRRQIGVVPQDTFLFS 427
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 MlTVEEHIwFYARlKGLSEKHVKAEMEQMALD---VGLPP---SKLKSKTSQLSGGmQR-KLSVALAFVGGSKVVILDEP 1059
Cdd:COG1132 428 G-TIRENI-RYGR-PDATDEEVEEAAKAAQAHefiEALPDgydTVVGERGVNLSGG-QRqRIAIARALLKDPPILILDEA 503
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1060 TAGVDPYSRRGIWELLLKYRQGRTIILSTH------HMdeadilgDRIAIISHGKLCCVGS 1114
Cdd:COG1132 504 TSALDTETEALIQEALERLMKGRTTIVIAHrlstirNA-------DRILVLDDGRIVEQGT 557
|
|
| LolD |
COG1136 |
ABC-type lipoprotein export system, ATPase component [Cell wall/membrane/envelope biogenesis]; |
1911-2123 |
1.11e-21 |
|
ABC-type lipoprotein export system, ATPase component [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440751 [Multi-domain] Cd Length: 227 Bit Score: 95.88 E-value: 1.11e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYRRK--RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDAFLNKNSI-------L 1980
Cdd:COG1136 2 SPLLELRNLTKSYGTGegEVTALRGVSLSIEAGEFVAIVGPSGSGKSTLLNILGGlDRP-TSGEVLIDGQDIsslsereL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1981 SNIHevHQNMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:COG1136 81 ARLR--RRHIGFVFQFFNLLPELTALENVALPLLLAGVSRKERRERARELLERVGLGDRLDHRPSQLSGGQQQRVAIARA 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSmEECEALCTRMAIMVNGR 2123
Cdd:COG1136 159 LVNRPKLILADEPTGNLDSKTGEEVLELLRELNRElGTTIVMVTHD-PELAARADRVIRLRDGR 221
|
|
| TauB |
COG4525 |
ABC-type taurine transport system, ATPase component [Inorganic ion transport and metabolism]; |
900-1105 |
1.25e-21 |
|
ABC-type taurine transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443596 [Multi-domain] Cd Length: 262 Bit Score: 96.85 E-value: 1.25e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVY--RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEmSSIRqnlGV 977
Cdd:COG4525 5 TVRHVSVRYpgGGQPQPALQDVSLTIESGEFVVALGASGCGKTTLLNLIAGFLAPSSGEITLDGVPVTGP-GADR---GV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIWFYARLKGLSekhvKAEMEQMALD----VGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:COG4525 81 VFQKDALLPWLNVLDNVAFGLRLRGVP----KAERRARAEEllalVGLADFA-RRRIWQLSGGMRQRVGIARALAADPRF 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIIS 1105
Cdd:COG4525 156 LLMDEPFGALDALTREQMQELLLDVwqRTGKGVFLITHSVEEALFLATRLVVMS 209
|
|
| ABCC_cytochrome_bd |
cd03247 |
ATP-binding cassette domain of CydCD, subfamily C; The CYD subfamily implicated in cytochrome ... |
899-1109 |
1.51e-21 |
|
ATP-binding cassette domain of CydCD, subfamily C; The CYD subfamily implicated in cytochrome bd biogenesis. The CydC and CydD proteins are important for the formation of cytochrome bd terminal oxidase of E. coli and it has been proposed that they were necessary for biosynthesis of the cytochrome bd quinol oxidase and for periplasmic c-type cytochromes. CydCD were proposed to determine a heterooligomeric complex important for heme export into the periplasm or to be involved in the maintenance of the proper redox state of the periplasmic space. In Bacillus subtilis, the absence of CydCD does not affect the presence of halo-cytochrome c in the membrane and this observation suggests that CydCD proteins are not involved in the export of heme in this organism.
Pssm-ID: 213214 [Multi-domain] Cd Length: 178 Bit Score: 93.92 E-value: 1.51e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC 978
Cdd:cd03247 1 LSINNVSFSYPEQEQQVLKNLSLELKQGEKIALLGRSGSGKSTLLQLLTGDLKPQQGEITLDGVPVSDLEKALSSLISVL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMlTVEEhiwfyarlkglsekhvkaemeqmalDVGLppsklksktsQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03247 81 NQRPYLFDT-TLRN-------------------------NLGR----------RFSGGERQRLALARILLQDAPIVLLDE 124
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMdeadiLG----DRIAIISHGKL 1109
Cdd:cd03247 125 PTVGLDPITERQLLSLIFEVLKDKTLIWITHHL-----TGiehmDKILFLENGKI 174
|
|
| nickel_nikD |
TIGR02770 |
nickel import ATP-binding protein NikD; This family represents the NikD subunit of a ... |
915-1109 |
1.55e-21 |
|
nickel import ATP-binding protein NikD; This family represents the NikD subunit of a multisubunit nickel import ABC transporter complex. Nickel, once imported, may be used in urease and in certain classes of hydrogenase and superoxide dismutase. NikD and NikE are homologous. [Transport and binding proteins, Cations and iron carrying compounds]
Pssm-ID: 131817 [Multi-domain] Cd Length: 230 Bit Score: 95.51 E-value: 1.55e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPP----TSGTAYILGKDIRSemSSIRQNLGVCPQHN---VLFDM 987
Cdd:TIGR02770 1 LVQDLNLSLKRGEVLALVGESGSGKSLTCLAILGLLPPgltqTSGEILLDGRPLLP--LSIRGRHIATIMQNprtAFNPL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 988 LTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSK--LKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:TIGR02770 79 FTMGNHAIETLRSLGKLSKQARALILEALEAVGLPDPEevLKKYPFQLSGGMLQRVMIALALLLEPPFLIADEPTTDLDV 158
|
170 180 190 200
....*....|....*....|....*....|....*....|....*.
gi 568925948 1066 YSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:TIGR02770 159 VNQARVLKLLRELRQlfGTGILLITHDLGVVARIADEVAVMDDGRI 204
|
|
| ABCG_White |
cd03234 |
White pigment protein homolog of ABCG transporter subfamily; The White subfamily represents ... |
1938-2104 |
1.56e-21 |
|
White pigment protein homolog of ABCG transporter subfamily; The White subfamily represents ABC transporters homologous to the Drosophila white gene, which acts as a dimeric importer for eye pigment precursors. The eye pigmentation of Drosophila is developed from the synthesis and deposition in the cells of red pigments, which are synthesized from guanine, and brown pigments, which are synthesized from tryptophan. The pigment precursors are encoded by the white, brown, and scarlet genes, respectively. Evidence from genetic and biochemical studies suggest that the White and Brown proteins function as heterodimers to import guanine, while the White and Scarlet proteins function to import tryptophan. However, a recent study also suggests that White may be involved in the transport of a metabolite, such as 3-hydroxykynurenine, across intracellular membranes. Mammalian ABC transporters belonging to the White subfamily (ABCG1, ABCG5, and ABCG8) have been shown to be involved in the regulation of lipid-trafficking mechanisms in macrophages, hepatocytes, and intestinal mucosa cells. ABCG1 (ABC8), the human homolog of the Drosophila white gene is induced in monocyte-derived macrophages during cholesterol influx mediated by acetylated low-density lipoprotein. It is possible that human ABCG1 forms heterodimers with several heterologous partners.
Pssm-ID: 213201 [Multi-domain] Cd Length: 226 Bit Score: 95.42 E-value: 1.56e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTG---DTPVTRGDAFLNKNSilSNIHEVHQNMGYCPQFDAITELLTGREHVEFFAL 2014
Cdd:cd03234 30 VESGQVMAILGSSGSGKTTLLDAISGrveGGGTTSGQILFNGQP--RKPDQFQKCVAYVRQDDILLPGLTVRETLTYTAI 107
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2015 LR-------GVPEKEVGKVGewaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPkarrFLwn 2087
Cdd:cd03234 108 LRlprkssdAIRKKRVEDVL---LRDLALTRIGGNLVKGISGGERRRVSIAVQLLWDPKVLILDEPTSGLDS----FT-- 178
|
170 180
....*....|....*....|....
gi 568925948 2088 cALSIV-------KEGRSVVLTSH 2104
Cdd:cd03234 179 -ALNLVstlsqlaRRNRIVILTIH 201
|
|
| PRK10895 |
PRK10895 |
lipopolysaccharide ABC transporter ATP-binding protein; Provisional |
902-1114 |
1.58e-21 |
|
lipopolysaccharide ABC transporter ATP-binding protein; Provisional
Pssm-ID: 182817 [Multi-domain] Cd Length: 241 Bit Score: 96.12 E-value: 1.58e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 902 QNLVKVYRdGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR--SEMSSIRQNLGVCP 979
Cdd:PRK10895 7 KNLAKAYK-GRRV-VEDVSLTVNSGEIVGLLGPNGAGKTTTFYMVVGIVPRDAGNIIIDDEDISllPLHARARRGIGYLP 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 980 QHNVLFDMLTVEEHIWFYARL-KGLSEKHVKAEMEQMALDVGLppSKLKSKTSQ-LSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:PRK10895 85 QEASIFRRLSVYDNLMAVLQIrDDLSAEQREDRANELMEEFHI--EHLRDSMGQsLSGGERRRVEIARALAANPKFILLD 162
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK10895 163 EPFAGVDPISVIDIKRIIEHLRdSGLGVLITDHNVRETLAVCERAYIVSQGHLIAHGT 220
|
|
| MlaF |
COG1127 |
ATPase subunit MlaF of the ABC-type intermembrane phospholipid transporter Mla [Cell wall ... |
1913-2135 |
1.80e-21 |
|
ATPase subunit MlaF of the ABC-type intermembrane phospholipid transporter Mla [Cell wall/membrane/envelope biogenesis];
Pssm-ID: 440744 [Multi-domain] Cd Length: 241 Bit Score: 95.82 E-value: 1.80e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKiyRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL----SNIHEVHQ 1988
Cdd:COG1127 5 MIEVRNLTK--SFGDRVVLDGVSLDVPRGEILAIIGGSGSGKSVLLKLIIGLLRPDSGEILVDGQDITglseKELYELRR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQ----FDAitelLTGREHVEFFalLR---GVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMAL 2061
Cdd:COG1127 83 RIGMLFQggalFDS----LTVFENVAFP--LRehtDLSEAEIRELVLEKLELVGLPGAADKMPSELSGGMRKRVALARAL 156
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2062 IGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:COG1127 157 ALDPEILLYDEPTAGLDPITSAVIDELIRELRDElGLTSVVVTHDLDSAFAIADRVAVLADGKIIAEGTPEELLA 231
|
|
| ThiQ |
COG3840 |
ABC-type thiamine transport system, ATPase component ThiQ [Coenzyme transport and metabolism]; |
926-1108 |
2.87e-21 |
|
ABC-type thiamine transport system, ATPase component ThiQ [Coenzyme transport and metabolism];
Pssm-ID: 443051 [Multi-domain] Cd Length: 232 Bit Score: 94.82 E-value: 2.87e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE------MSSIRQnlgvcpQHNvLFDMLTVEEHIWF--Y 997
Cdd:COG3840 25 GERVAILGPSGAGKSTLLNLIAGFLPPDSGRILWNGQDLTALppaerpVSMLFQ------ENN-LFPHLTVAQNIGLglR 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 998 ARLKgLSEKHvKAEMEQMALDVGLPpSKLKSKTSQLSGG-MQRklsVALA--FVGGSKVVILDEPTAGVDPYSRRGIWEL 1074
Cdd:COG3840 98 PGLK-LTAEQ-RAQVEQALERVGLA-GLLDRLPGQLSGGqRQR---VALArcLVRKRPILLLDEPFSALDPALRQEMLDL 171
|
170 180 190
....*....|....*....|....*....|....*.
gi 568925948 1075 L--LKYRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:COG3840 172 VdeLCRERGLTVLMVTHDPEDAARIADRVLLVADGR 207
|
|
| ABC_Org_Solvent_Resistant |
cd03261 |
ATP-binding cassette transport system involved in resistance to organic solvents; ABC ... |
1914-2135 |
3.25e-21 |
|
ATP-binding cassette transport system involved in resistance to organic solvents; ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213228 [Multi-domain] Cd Length: 235 Bit Score: 94.88 E-value: 3.25e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKiyRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNI----HEVHQN 1989
Cdd:cd03261 1 IELRGLTK--SFGGRTVLKGVDLDVRRGEILAIIGPSGSGKSTLLRLIVGLLRPDSGEVLIDGEDISGLSeaelYRLRRR 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQFDAITELLTGREHVEFFalLR---GVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPP 2066
Cdd:cd03261 79 MGMLFQSGALFDSLTVFENVAFP--LRehtRLSEEEIREIVLEKLEAVGLRGAEDLYPAELSGGMKKRVALARALALDPE 156
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2067 VVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:cd03261 157 LLLYDEPTAGLDPIASGVIDDLIRSLKKElGLTSIMVTHDLDTAFAIADRIAVLYDGKIVAEGTPEELRA 226
|
|
| ABC_KpsT_Wzt |
cd03220 |
ATP-binding cassette component of polysaccharide transport system; The KpsT/Wzt ABC ... |
893-1109 |
3.82e-21 |
|
ATP-binding cassette component of polysaccharide transport system; The KpsT/Wzt ABC transporter subfamily is involved in extracellular polysaccharide export. Among the variety of membrane-linked or extracellular polysaccharides excreted by bacteria, only capsular polysaccharides, lipopolysaccharides, and teichoic acids have been shown to be exported by ABC transporters. A typical system is made of a conserved integral membrane and an ABC. In addition to these proteins, capsular polysaccharide exporter systems require two 'accessory' proteins to perform their function: a periplasmic (E.coli) or a lipid-anchored outer membrane protein called OMA (Neisseria meningitidis and Haemophilus influenza) and a cytoplasmic membrane protein MPA2.
Pssm-ID: 213187 [Multi-domain] Cd Length: 224 Bit Score: 94.14 E-value: 3.82e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 893 THLRLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKdirseMSSIr 972
Cdd:cd03220 15 GGSSSLKKLGILGRKGEVGEFWALKDVSFEVPRGERIGLIGRNGAGKSTLLRLLAGIYPPDSGTVTVRGR-----VSSL- 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 qnLGVcpqhNVLFD-MLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSkLKSKTSQLSGGMQRKL--SVALAFvg 1049
Cdd:cd03220 89 --LGL----GGGFNpELTGRENIYLNGRLLGLSRKEIDEKIDEIIEFSELGDF-IDLPVKTYSSGMKARLafAIATAL-- 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1050 GSKVVILDEPTAGVDPYSRRGIWELLL-KYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03220 160 EPDILLIDEVLAVGDAAFQEKCQRRLReLLKQGKTVILVSHDPSSIKRLCDRALVLEKGKI 220
|
|
| ABC_KpsT_Wzt |
cd03220 |
ATP-binding cassette component of polysaccharide transport system; The KpsT/Wzt ABC ... |
1905-2128 |
4.90e-21 |
|
ATP-binding cassette component of polysaccharide transport system; The KpsT/Wzt ABC transporter subfamily is involved in extracellular polysaccharide export. Among the variety of membrane-linked or extracellular polysaccharides excreted by bacteria, only capsular polysaccharides, lipopolysaccharides, and teichoic acids have been shown to be exported by ABC transporters. A typical system is made of a conserved integral membrane and an ABC. In addition to these proteins, capsular polysaccharide exporter systems require two 'accessory' proteins to perform their function: a periplasmic (E.coli) or a lipid-anchored outer membrane protein called OMA (Neisseria meningitidis and Haemophilus influenza) and a cytoplasmic membrane protein MPA2.
Pssm-ID: 213187 [Multi-domain] Cd Length: 224 Bit Score: 94.14 E-value: 4.90e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1905 LDGGGQNDILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsiLSNIH 1984
Cdd:cd03220 12 TYKGGSSSLKKLGILGRKGEVGEFWALKDVSFEVPRGERIGLIGRNGAGKSTLLRLLAGIYPPDSGTVTVRGR--VSSLL 89
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVhqNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGG 2064
Cdd:cd03220 90 GL--GGGFNPE-------LTGRENIYLNGRLLGLSRKEIDEKIDEIIEFSELGDFIDLPVKTYSSGMKARLAFAIATALE 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2065 PPVVFLDEPTTGMDP----KARRFLwncaLSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:cd03220 161 PDILLIDEVLAVGDAafqeKCQRRL----RELLKQGKTVILVSHDPSSIKRLCDRALVLEKGKIRFDG 224
|
|
| CydC |
TIGR02868 |
thiol reductant ABC exporter, CydC subunit; The gene pair cydCD encodes an ABC-family ... |
905-1090 |
7.58e-21 |
|
thiol reductant ABC exporter, CydC subunit; The gene pair cydCD encodes an ABC-family transporter in which each gene contains an N-terminal membrane-spanning domain (pfam00664) and a C-terminal ATP-binding domain (pfam00005). In E. coli these genes were discovered as mutants which caused the terminal heme-copper oxidase complex cytochrome bd to fail to assemble. Recent work has shown that the transporter is involved in export of redox-active thiol compounds such as cysteine and glutathione. The linkage to assembly of the cytochrome bd complex is further supported by the conserved operon structure found outside the gammaproteobacteria (cydABCD) containing both the transporter and oxidase genes components. The genes used as the seed members for this model are all either found in the gammproteobacterial context or the CydABCD context. All members of this family scoring above trusted at the time of its creation were from genomes which encode a cytochrome bd complex.
Pssm-ID: 274331 [Multi-domain] Cd Length: 530 Bit Score: 98.97 E-value: 7.58e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 905 VKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNV 983
Cdd:TIGR02868 340 LSAGYPGAPPVLDGVSLDLPPGERVAILGPSGSGKSTLLATLAGLLDPLQGEVTLDGVPVSSlDQDEVRRRVSVCAQDAH 419
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 984 LFDMlTVEEHIWFyARlKGLSEkhvkAEMEQMALDVGLPP----------SKLKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:TIGR02868 420 LFDT-TVRENLRL-AR-PDATD----EELWAALERVGLADwlralpdgldTVLGEGGARLSGGERQRLALARALLADAPI 492
|
170 180 190
....*....|....*....|....*....|....*..
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHH 1090
Cdd:TIGR02868 493 LLLDEPTEHLDAETADELLEDLLAALSGRTVVLITHH 529
|
|
| PotA |
COG3842 |
ABC-type Fe3+/spermidine/putrescine transport systems, ATPase component [Amino acid transport ... |
1913-2170 |
7.85e-21 |
|
ABC-type Fe3+/spermidine/putrescine transport systems, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 443052 [Multi-domain] Cd Length: 353 Bit Score: 96.71 E-value: 7.85e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIhEVHQ-NMG 1991
Cdd:COG3842 5 ALELENVSKRY--GDVTALDDVSLSIEPGEFVALLGPSGCGKTTLLRMIAGFETPDSGRILLDGRDV-TGL-PPEKrNVG 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDAiteL---LTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:COG3842 81 MVFQDYA---LfphLTVAENVAFGLRMRGVPKAEIRARVAELLELVGLEGLADRYPHQLSGGQQQRVALARALAPEPRVL 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL----KNRFgdgyti 2143
Cdd:COG3842 158 LLDEPLSALDAKLREEMREELRRLQRElGITFIYVTHDQEEALALADRIAVMNDGRIEQVGTPEEIyerpATRF------ 231
|
250 260
....*....|....*....|....*....
gi 568925948 2144 vvriagsnpdlkpVQEFFGLA--FPGSVL 2170
Cdd:COG3842 232 -------------VADFIGEAnlLPGTVL 247
|
|
| AztA |
NF040873 |
zinc ABC transporter ATP-binding protein AztA; |
914-1094 |
8.06e-21 |
|
zinc ABC transporter ATP-binding protein AztA;
Pssm-ID: 468810 [Multi-domain] Cd Length: 191 Bit Score: 92.30 E-value: 8.06e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 914 VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayilgkdiRSEMSSIRqnLGVCPQHNVLFDML--TVE 991
Cdd:NF040873 6 PVLHGVDLTIPAGSLTAVVGPNGSGKSTLLKVLAGVLRPTSGT--------VRRAGGAR--VAYVPQRSEVPDSLplTVR 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 992 E--------HIWFYARLKGLSEKHVKAEMEQMALDvGLPPSKLksktSQLSGG-MQRKLsVALAFVGGSKVVILDEPTAG 1062
Cdd:NF040873 76 DlvamgrwaRRGLWRRLTRDDRAAVDDALERVGLA-DLAGRQL----GELSGGqRQRAL-LAQGLAQEADLLLLDEPTTG 149
|
170 180 190
....*....|....*....|....*....|...
gi 568925948 1063 VDPYSRRGIWELLLKY-RQGRTIILSTHHMDEA 1094
Cdd:NF040873 150 LDAESRERIIALLAEEhARGATVVVVTHDLELV 182
|
|
| cbiO |
PRK13652 |
cobalt transporter ATP-binding subunit; Provisional |
899-1130 |
8.76e-21 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 172200 [Multi-domain] Cd Length: 277 Bit Score: 94.87 E-value: 8.76e-21
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRdGMKVAVDGLalNFYEGQIT--SFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNL 975
Cdd:PRK13652 4 IETRDLCYSYS-GSKEALNNI--NFIAPRNSriAVIGPNGAGKSTLFRHFNGILKPTSGSVLIRGEPITKEnIREVRKFV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQH--NVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLppSKLKSKTSQ-LSGGMQRKLSVALAFVGGSK 1052
Cdd:PRK13652 81 GLVFQNpdDQIFSP-TVEQDIAFGPINLGLDEETVAHRVSSALHMLGL--EELRDRVPHhLSGGEKKRVAIAGVIAMEPQ 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS--SLFLKNQLGTGYYL 1128
Cdd:PRK13652 158 VLVLDEPTAGLDPQGVKELIDFLndLPETYGMTVIFSTHQLDLVPEMADYIYVMDKGRIVAYGTveEIFLQPDLLARVHL 237
|
..
gi 568925948 1129 TL 1130
Cdd:PRK13652 238 DL 239
|
|
| Uup |
COG0488 |
ATPase components of ABC transporters with duplicated ATPase domains [General function ... |
896-1109 |
1.06e-20 |
|
ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only];
Pssm-ID: 440254 [Multi-domain] Cd Length: 520 Bit Score: 98.60 E-value: 1.06e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 896 RLG---VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTaYILGKDIRsemssir 972
Cdd:COG0488 310 RLGkkvLELEGLSKSYGD--KTLLDDLSLRIDRGDRIGLIGPNGAGKSTLLKLLAGELEPDSGT-VKLGETVK------- 379
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 qnLGVCPQHNVLFDM-LTVEEHIWFYARlkGLSEKHVKAEMEQMaldvGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:COG0488 380 --IGYFDQHQEELDPdKTVLDELRDGAP--GGTEQEVRGYLGRF----LFSGDDAFKPVGVLSGGEKARLALAKLLLSPP 451
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYrQGrTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG0488 452 NVLLLDEPTNHLDIETLEALEEALDDF-PG-TVLLVSHDRYFLDRVATRILEFEDGGV 507
|
|
| cbiO |
PRK13643 |
energy-coupling factor transporter ATPase; |
921-1114 |
1.09e-20 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184203 [Multi-domain] Cd Length: 288 Bit Score: 94.80 E-value: 1.09e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTA-----YILGKDIRSEMSSIRQNLGVCPQ--HNVLFDMlTVEEH 993
Cdd:PRK13643 27 LEVKKGSYTALIGHTGSGKSTLLQHLNGLLQPTEGKVtvgdiVVSSTSKQKEIKPVRKKVGVVFQfpESQLFEE-TVLKD 105
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 994 IWFYARLKGLSekhvKAEMEQMALD----VGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRR 1069
Cdd:PRK13643 106 VAFGPQNFGIP----KEKAEKIAAEklemVGLADEFWEKSPFELSGGQMRRVAIAGILAMEPEVLVLDEPTAGLDPKARI 181
|
170 180 190 200
....*....|....*....|....*....|....*....|....*.
gi 568925948 1070 GIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13643 182 EMMQLFESIHQsGQTVVLVTHLMDDVADYADYVYLLEKGHIISCGT 227
|
|
| ModF |
COG1119 |
ABC-type molybdenum transport system, ATPase component ModF/photorepair protein PhrA ... |
1911-2123 |
1.15e-20 |
|
ABC-type molybdenum transport system, ATPase component ModF/photorepair protein PhrA [Inorganic ion transport and metabolism];
Pssm-ID: 440736 [Multi-domain] Cd Length: 250 Bit Score: 93.61 E-value: 1.15e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFlnknSIL------SNIH 1984
Cdd:COG1119 1 DPLLELRNVT--VRRGGKTILDDISWTVKPGEHWAILGPNGAGKSTLLSLITGDLPPTYGNDV----RLFgerrggEDVW 74
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVHQNMGYC-PQF-DAITELLTGREHVE--FFA---LLRGVPEKEVGKVGEWaIRKLGLVKYGEKYASNYSGGNKRKLST 2057
Cdd:COG1119 75 ELRKRIGLVsPALqLRFPRDETVLDVVLsgFFDsigLYREPTDEQRERAREL-LELLGLAHLADRPFGTLSQGEQRRVLI 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFLWNcALS-IVKEG-RSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG1119 154 ARALVKDPELLILDEPTAGLDLGARELLLA-LLDkLAAEGaPTLVLVTHHVEEIPPGITHVLLLKDGR 220
|
|
| potA |
PRK09452 |
spermidine/putrescine ABC transporter ATP-binding protein PotA; |
899-1114 |
1.47e-20 |
|
spermidine/putrescine ABC transporter ATP-binding protein PotA;
Pssm-ID: 236523 [Multi-domain] Cd Length: 375 Bit Score: 96.17 E-value: 1.47e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:PRK09452 15 VELRGISKSFDG--KEVISNLDLTINNGEFLTLLGPSGCGKTTVLRLIAGFETPDSGRIMLDGQDI-THVPAENRHVNTV 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKA---------EMEQMAldvglppsklKSKTSQLSGGMQRKLSVALAFVG 1049
Cdd:PRK09452 92 FQSYALFPHMTVFENVAFGLRMQKTPAAEITPrvmealrmvQLEEFA----------QRKPHQLSGGQQQRVAIARAVVN 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1050 GSKVVILDEPTAGVDpYSRRGIWELLLKYRQ---GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK09452 162 KPKVLLLDESLSALD-YKLRKQMQNELKALQrklGITFVFVTHDQEEALTMSDRIVVMRDGRIEQDGT 228
|
|
| met_CoM_red_A2 |
TIGR03269 |
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in ... |
883-1113 |
1.50e-20 |
|
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in methanogenesis, methyl coenzyme M reductase, contains alpha, beta, and gamma chains. In older literature, the complex of alpha, beta, and gamma chains was termed component C, while this single chain protein was termed methyl coenzyme M reductase system component A2. [Energy metabolism, Methanogenesis]
Pssm-ID: 132313 [Multi-domain] Cd Length: 520 Bit Score: 97.95 E-value: 1.50e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 883 VSEICME-----EEPTHLRLG---VSIQNLVKVY---RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP 951
Cdd:TIGR03269 256 VVAVFMEgvsevEKECEVEVGepiIKVRNVSKRYisvDRGVVKAVDNVSLEVKEGEIFGIVGTSGAGKTTLSKIIAGVLE 335
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 952 PTSGTAYILGKDIRSEMSSIR--------QNLGVCPQ------HNVLFDMLTVEEHIWF---YARLK--------GLSEK 1006
Cdd:TIGR03269 336 PTSGEVNVRVGDEWVDMTKPGpdgrgrakRYIGILHQeydlypHRTVLDNLTEAIGLELpdeLARMKavitlkmvGFDEE 415
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1007 HVKAEMEQMaldvglppsklkskTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTI 1084
Cdd:TIGR03269 416 KAEEILDKY--------------PDELSEGERHRVALAQVLIKEPRIVILDEPTGTMDPITKVDVTHSILKAREemEQTF 481
|
250 260
....*....|....*....|....*....
gi 568925948 1085 ILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:TIGR03269 482 IIVSHDMDFVLDVCDRAALMRDGKIVKIG 510
|
|
| MglA |
COG1129 |
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism]; |
915-1109 |
2.00e-20 |
|
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440745 [Multi-domain] Cd Length: 497 Bit Score: 97.40 E-value: 2.00e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK--DIRSEMSSIRQNLGVCP---QHNVLFDMLT 989
Cdd:COG1129 267 VVRDVSFSVRAGEILGIAGLVGAGRTELARALFGADPADSGEIRLDGKpvRIRSPRDAIRAGIAYVPedrKGEGLVLDLS 346
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 990 VEEHIWF-----YARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKlsVALA--FVGGSKVVILDEPTAG 1062
Cdd:COG1129 347 IRENITLasldrLSRGGLLDRRRERALAEEYIKRLRIKTPSPEQPVGNLSGGNQQK--VVLAkwLATDPKVLILDEPTRG 424
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 1063 VDPYSRRGIWELLLKY-RQGRTIILSTHHMDEadILG--DRIAIISHGKL 1109
Cdd:COG1129 425 IDVGAKAEIYRLIRELaAEGKAVIVISSELPE--LLGlsDRILVMREGRI 472
|
|
| CydC |
COG4987 |
ABC-type transport system involved in cytochrome bd biosynthesis, fused ATPase and permease ... |
1914-2138 |
2.55e-20 |
|
ABC-type transport system involved in cytochrome bd biosynthesis, fused ATPase and permease components [Energy production and conversion, Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 444011 [Multi-domain] Cd Length: 569 Bit Score: 97.53 E-value: 2.55e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:COG4987 334 LELEDVSFRYPGAGRPVLDGLSLTLPPGERVAIVGPSGSGKSTLLALLLRFLDPQSGSITLGGVDLRDlDEDDLRRRIAV 413
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQ----FDA-ITE-LLTGREHV---EFFALLRGVpekevgKVGEWaIRKL--GL---VkyGEkYASNYSGGNKRKLSTA 2058
Cdd:COG4987 414 VPQrphlFDTtLREnLRLARPDAtdeELWAALERV------GLGDW-LAALpdGLdtwL--GE-GGRRLSGGERRRLALA 483
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2059 MALIGGPPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSHSMEECEAlCTRMAIMVNGRFRCLGSVQHLKNRFG 2138
Cdd:COG4987 484 RALLRDAPILLLDEPTEGLDAATEQALLA-DLLEALAGRTVLLITHRLAGLER-MDRILVLEDGRIVEQGTHEELLAQNG 561
|
|
| livG |
PRK11300 |
leucine/isoleucine/valine transporter ATP-binding subunit; Provisional |
909-1108 |
3.02e-20 |
|
leucine/isoleucine/valine transporter ATP-binding subunit; Provisional
Pssm-ID: 183080 [Multi-domain] Cd Length: 255 Bit Score: 92.75 E-value: 3.02e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCP--QHNVLFD 986
Cdd:PRK11300 14 RFGGLLAVNNVNLEVREQEIVSLIGPNGAGKTTVFNCLTGFYKPTGGTILLRGQHIEGLPGHQIARMGVVRtfQHVRLFR 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 MLTVEE------HIWFYAR-LKGL--SEKHVKAEMEQMA-----LD-VGLPPSKLKSkTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:PRK11300 94 EMTVIEnllvaqHQQLKTGlFSGLlkTPAFRRAESEALDraatwLErVGLLEHANRQ-AGNLAYGQQRRLEIARCMVTQP 172
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK11300 173 EILMLDEPAAGLNPKETKELDELIAELRNehNVTVLLIEHDMKLVMGISDRIYVVNQGT 231
|
|
| cbiO |
PRK13645 |
energy-coupling factor transporter ATPase; |
915-1114 |
6.54e-20 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184204 [Multi-domain] Cd Length: 289 Bit Score: 92.38 E-value: 6.54e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTA----YILGKDIRS--EMSSIRQNLGVC---PQHNVLF 985
Cdd:PRK13645 26 ALNNTSLTFKKNKVTCVIGTTGSGKSTMIQLTNGLIISETGQTivgdYAIPANLKKikEVKRLRKEIGLVfqfPEYQLFQ 105
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 DmlTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:PRK13645 106 E--TIEKDIAFGPVNLGENKQEAYKKVPELLKLVQLPEDYVKRSPFELSGGQKRRVALAGIIAMDGNTLVLDEPTGGLDP 183
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1066 YSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13645 184 KGEEDFINLFERLnkEYKKRIIMVTHNMDQVLRIADEVIVMHEGKVISIGS 234
|
|
| ABC_TM1139_LivF_branched |
cd03224 |
ATP-binding cassette domain of branched-chain amino acid transporter; LivF (TM1139) is part of ... |
1914-2125 |
7.99e-20 |
|
ATP-binding cassette domain of branched-chain amino acid transporter; LivF (TM1139) is part of the LIV-I bacterial ABC-type two-component transport system that imports neutral, branched-chain amino acids. The E. coli branched-chain amino acid transporter comprises a heterodimer of ABC transporters (LivF and LivG), a heterodimer of six-helix TM domains (LivM and LivH), and one of two alternative soluble periplasmic substrate binding proteins (LivK or LivJ). ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules.
Pssm-ID: 213191 [Multi-domain] Cd Length: 222 Bit Score: 90.57 E-value: 7.99e-20
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHE-VHQNMG 1991
Cdd:cd03224 1 LEVENLNAGY--GKSQILFGVSLTVPEGEIVALLGRNGAGKTTLLKTIMGLLPPRSGSIRFDGRDITGlPPHErARAGIG 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDAITELLTGREHVEFFALLRGvpekevGKVGEWAIRKLglvkYG---------EKYASNYSGGNKRKLSTAMALI 2062
Cdd:cd03224 79 YVPEGRRIFPELTVEENLLLGAYARR------RAKRKARLERV----YElfprlkerrKQLAGTLSGGEQQMLAIARALM 148
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2063 GGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFR 2125
Cdd:cd03224 149 SRPKLLLLDEPSEGLAPKIVEEIFEAIRELRDEGVTILLVEQNARFALEIADRAYVLERGRVV 211
|
|
| PRK13539 |
PRK13539 |
cytochrome c biogenesis protein CcmA; Provisional |
912-1089 |
1.02e-19 |
|
cytochrome c biogenesis protein CcmA; Provisional
Pssm-ID: 237421 [Multi-domain] Cd Length: 207 Bit Score: 89.55 E-value: 1.02e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 912 MKVAVDGLA-----------LNFY--EGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayILGKDIRSEMSSIRQN---L 975
Cdd:PRK13539 1 MMLEGEDLAcvrggrvlfsgLSFTlaAGEALVLTGPNGSGKTTLLRLIAGLLPPAAGT--IKLDGGDIDDPDVAEAchyL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GvcpQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEqmalDVGLPP-SKLKSKTsqLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:PRK13539 79 G---HRNAMKPALTVAENLEFWAAFLGGEELDIAAALE----AVGLAPlAHLPFGY--LSAGQKRRVALARLLVSNRPIW 149
|
170 180 190
....*....|....*....|....*....|....*.
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTH 1089
Cdd:PRK13539 150 ILDEPTAALDAAAVALFAELIRAHLaQGGIVIAATH 185
|
|
| TagH |
COG1134 |
ABC-type polysaccharide/polyol phosphate transport system, ATPase component [Carbohydrate ... |
1911-2130 |
1.10e-19 |
|
ABC-type polysaccharide/polyol phosphate transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440749 [Multi-domain] Cd Length: 245 Bit Score: 90.53 E-value: 1.10e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYR--------------------RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRG 1970
Cdd:COG1134 2 SSMIEVENVSKSYRlyhepsrslkelllrrrrtrREEFWALKDVSFEVERGESVGIIGRNGAGKSTLLKLIAGILEPTSG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1971 DAFLNKN--SILsnihEVhqNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGK----VGEWAirklGLVKYGEKYA 2044
Cdd:COG1134 82 RVEVNGRvsALL----EL--GAGFHPE-------LTGRENIYLNGRLLGLSRKEIDEkfdeIVEFA----ELGDFIDQPV 144
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2045 SNYSGGNKRKL--STAMALigGPPVVFLDEPTTGMDP----KARRFLwncaLSIVKEGRSVVLTSHSMEECEALCTRMAI 2118
Cdd:COG1134 145 KTYSSGMRARLafAVATAV--DPDILLVDEVLAVGDAafqkKCLARI----RELRESGRTVIFVSHSMGAVRRLCDRAIW 218
|
250
....*....|..
gi 568925948 2119 MVNGRFRCLGSV 2130
Cdd:COG1134 219 LEKGRLVMDGDP 230
|
|
| ABCC_bacteriocin_exporters |
cd03245 |
ATP-binding cassette domain of bacteriocin exporters, subfamily C; Many non-lantibiotic ... |
1914-2123 |
1.23e-19 |
|
ATP-binding cassette domain of bacteriocin exporters, subfamily C; Many non-lantibiotic bacteriocins of lactic acid bacteria are produced as precursors which have N-terminal leader peptides that share similarities in amino acid sequence and contain a conserved processing site of two glycine residues in positions -1 and -2. A dedicated ATP-binding cassette (ABC) transporter is responsible for the proteolytic cleavage of the leader peptides and subsequent translocation of the bacteriocins across the cytoplasmic membrane.
Pssm-ID: 213212 [Multi-domain] Cd Length: 220 Bit Score: 89.96 E-value: 1.23e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIH--EVHQNMG 1991
Cdd:cd03245 3 IEFRNVSFSYPNQEIPALDNVSLTIRAGEKVAIIGRVGSGKSTLLKLLAGLYKPTSGSVLLDGTDI-RQLDpaDLRRNIG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQ-----FDAITELLT-GREHVEFFALLRGVpekEVGKVGEWAIR-KLGL-VKYGEKyASNYSGGNKRKLSTAMALIG 2063
Cdd:cd03245 82 YVPQdvtlfYGTLRDNITlGAPLADDERILRAA---ELAGVTDFVNKhPNGLdLQIGER-GRGLSGGQRQAVALARALLN 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2064 GPPVVFLDEPTTGMDPKA-RRFLWNcaLSIVKEGRSVVLTSH--SMEEceaLCTRMAIMVNGR 2123
Cdd:cd03245 158 DPPILLLDEPTSAMDMNSeERLKER--LRQLLGDKTLIIITHrpSLLD---LVDRIIVMDSGR 215
|
|
| ccmA |
TIGR01189 |
heme ABC exporter, ATP-binding protein CcmA; This model describes the cyt c biogenesis protein ... |
909-1090 |
1.63e-19 |
|
heme ABC exporter, ATP-binding protein CcmA; This model describes the cyt c biogenesis protein encoded by ccmA in bacteria. An exception is, an arabidopsis protein. Quite likely this is encoded by an organelle. Bacterial c-type cytocromes are located on the periplasmic side of the cytoplasmic membrane. Several gene products encoded in a locus designated as 'ccm' are implicated in the transport and assembly of the functional cytochrome C. This cluster includes genes: ccmA;B;C;D;E;F;G and H. The posttranslational pathway includes the transport of heme moiety, the secretion of the apoprotein and the covalent attachment of the heme with the apoprotein. The proteins ccmA and B represent an ABC transporter; ccmC and D participate in heme transfer to ccmE, which function as a periplasmic heme chaperone. The presence of ccmF, G and H is suggested to be obligatory for the final functional assembly of cytochrome c. [Protein fate, Protein and peptide secretion and trafficking, Transport and binding proteins, Other]
Pssm-ID: 273491 [Multi-domain] Cd Length: 198 Bit Score: 88.95 E-value: 1.63e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDML 988
Cdd:TIGR01189 10 RGE-RMLFEGLSFTLNAGEALQVTGPNGIGKTTLLRILAGLLRPDSGEVRWNGTPLAEQRDEPHENILYLGHLPGLKPEL 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWFYARLKGLSEKHVKAEMEQMALD--VGLPpsklkskTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPY 1066
Cdd:TIGR01189 89 SALENLHFWAAIHGGAQRTIEDALAAVGLTgfEDLP-------AAQLSAGQQRRLALARLWLSRRPLWILDEPTTALDKA 161
|
170 180
....*....|....*....|....*
gi 568925948 1067 SRRGIWELLLKY-RQGRTIILSTHH 1090
Cdd:TIGR01189 162 GVALLAGLLRAHlARGGIVLLTTHQ 186
|
|
| PRK10070 |
PRK10070 |
proline/glycine betaine ABC transporter ATP-binding protein ProV; |
909-1132 |
1.96e-19 |
|
proline/glycine betaine ABC transporter ATP-binding protein ProV;
Pssm-ID: 182221 [Multi-domain] Cd Length: 400 Bit Score: 93.17 E-value: 1.96e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR----SEMSSI-RQNLGVCPQHNV 983
Cdd:PRK10070 37 KTGLSLGVKDASLAIEEGEIFVIMGLSGSGKSTMVRLLNRLIEPTRGQVLIDGVDIAkisdAELREVrRKKIAMVFQSFA 116
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 984 LFDMLTVEEHIWFYARLKGLSekhvKAEMEQMALD----VGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEP 1059
Cdd:PRK10070 117 LMPHMTVLDNTAFGMELAGIN----AEERREKALDalrqVGL-ENYAHSYPDELSGGMRQRVGLARALAINPDILLMDEA 191
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1060 TAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQLGTGYYLTLVK 1132
Cdd:PRK10070 192 FSALDPLIRTEMQDELVKLqaKHQRTIVFISHDLDEAMRIGDRIAIMQNGEVVQVGTPDEILNNPANDYVRTFFR 266
|
|
| ABCC_ATM1_transporter |
cd03253 |
ATP-binding cassette domain of iron-sulfur clusters transporter, subfamily C; ATM1 is an ABC ... |
908-1109 |
3.06e-19 |
|
ATP-binding cassette domain of iron-sulfur clusters transporter, subfamily C; ATM1 is an ABC transporter that is expressed in the mitochondria. Although the specific function of ATM1 is unknown, its disruption results in the accumulation of excess mitochondrial iron, loss of mitochondrial cytochromes, oxidative damage to mitochondrial DNA, and decreased levels of cytosolic heme proteins. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213220 [Multi-domain] Cd Length: 236 Bit Score: 89.21 E-value: 3.06e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFD 986
Cdd:cd03253 10 YDPGRPV-LKDVSFTIPAGKKVAIVGPSGSGKSTILRLLFRFYDVSSGSILIDGQDIREvTLDSLRRAIGVVPQDTVLFN 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 MlTVEEHIwFYARLKGLSEKHVKAEMEQMALD--VGLP---PSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTA 1061
Cdd:cd03253 89 D-TIGYNI-RYGRPDATDEEVIEAAKAAQIHDkiMRFPdgyDTIVGERGLKLSGGEKQRVAIARAILKNPPILLLDEATS 166
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1062 GVDPYSRRGIWELLLKYRQGRTIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:cd03253 167 ALDTHTEREIQAALRDVSKGRTTIVIAHRLSTI-VNADKIIVLKDGRI 213
|
|
| PRK13538 |
PRK13538 |
cytochrome c biogenesis heme-transporting ATPase CcmA; |
917-1090 |
3.32e-19 |
|
cytochrome c biogenesis heme-transporting ATPase CcmA;
Pssm-ID: 184125 [Multi-domain] Cd Length: 204 Bit Score: 87.94 E-value: 3.32e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 917 DGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWF 996
Cdd:PRK13538 18 SGLSFTLNAGELVQIEGPNGAGKTSLLRILAGLARPDAGEVLWQGEPIRRQRDEYHQDLLYLGHQPGIKTELTALENLRF 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 997 YARLKGLsekhVKAEMEQMALD-VGL------PpsklkskTSQLSGGMQRKlsVALA--FVGGSKVVILDEP-----TAG 1062
Cdd:PRK13538 98 YQRLHGP----GDDEALWEALAqVGLagfedvP-------VRQLSAGQQRR--VALArlWLTRAPLWILDEPftaidKQG 164
|
170 180
....*....|....*....|....*...
gi 568925948 1063 VDPYSRRgiweLLLKYRQGRTIILSTHH 1090
Cdd:PRK13538 165 VARLEAL----LAQHAEQGGMVILTTHQ 188
|
|
| cbiO |
PRK13636 |
cobalt transporter ATP-binding subunit; Provisional |
1910-2123 |
5.12e-19 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184196 [Multi-domain] Cd Length: 283 Bit Score: 89.52 E-value: 5.12e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI---LSNIHEV 1986
Cdd:PRK13636 2 EDYILKVEELNYNYSDGTH-ALKGININIKKGEVTAILGGNGAGKSTLFQNLNGILKPSSGRILFDGKPIdysRKGLMKL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNMGYCPQfDAITELLTGR--EHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGG 2064
Cdd:PRK13636 81 RESVGMVFQ-DPDNQLFSASvyQDVSFGAVNLKLPEDEVRKRVDNALKRTGIEHLKDKPTHCLSFGQKKRVAIAGVLVME 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2065 PPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13636 160 PKVLVLDEPTAGLDPMGVSEIMKLLVEMQKElGLTIIIATHDIDIVPLYCDNVFVMKEGR 219
|
|
| ABC_Carb_Monos_II |
cd03215 |
Second domain of the ATP-binding cassette component of monosaccharide transport system; This ... |
1913-2124 |
5.16e-19 |
|
Second domain of the ATP-binding cassette component of monosaccharide transport system; This family represents domain II of the carbohydrate uptake proteins that transport only monosaccharides (Monos). The Carb_Monos family is involved in the uptake of monosaccharides, such as pentoses (such as xylose, arabinose, and ribose) and hexoses (such as xylose, arabinose, and ribose), that cannot be broken down to simple sugars by hydrolysis. In members of Carb_Monos family the single hydrophobic gene product forms a homodimer, while the ABC protein represents a fusion of two nucleotide-binding domains. However, it is assumed that two copies of the ABC domains are present in the assembled transporter.
Pssm-ID: 213182 [Multi-domain] Cd Length: 182 Bit Score: 86.72 E-value: 5.16e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTkiyrrkRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHEVHQN-M 1990
Cdd:cd03215 4 VLEVRGLS------VKGAVRDVSFEVRAGEIVGIAGLVGNGQTELAEALFGLRPPASGEITLDGKPVtRRSPRDAIRAgI 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYcpqfdaitelltgrehveffallrgVPE--KEVGKVGEWAIRK-LGLvkygekyASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:cd03215 78 AY-------------------------VPEdrKREGLVLDLSVAEnIAL-------SSLLSGGNQQKVVLARWLARDPRV 125
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:cd03215 126 LILDEPTRGVDVGAKAEIYRLIRELADAGKAVLLISSELDELLGLCDRILVMYEGRI 182
|
|
| ABC_Class3 |
cd03229 |
ATP-binding cassette domain of the binding protein-dependent transport systems; This class is ... |
1914-2123 |
5.17e-19 |
|
ATP-binding cassette domain of the binding protein-dependent transport systems; This class is comprised of all BPD (Binding Protein Dependent) systems that are largely represented in archaea and eubacteria and are primarily involved in scavenging solutes from the environment. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213196 [Multi-domain] Cd Length: 178 Bit Score: 86.86 E-value: 5.17e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN---KNSILSNIHEVHQNM 1990
Cdd:cd03229 1 LELKNVSKRY--GQKTVLNDVSLNIEAGEIVALLGPSGSGKSTLLRCIAGLEEPDSGSILIDgedLTDLEDELPPLRRRI 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAITELLTGREHVEFfallrgvpekevgkvgewairklGLvkygekyasnySGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:cd03229 79 GMVFQDFALFPHLTVLENIAL-----------------------GL-----------SGGQQQRVALARALAMDPDVLLL 124
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03229 125 DEPTSALDPITRREVRALLKSLQAQlGITVVLVTHDLDEAARLADRVVVLRDGK 178
|
|
| cbiO |
PRK13640 |
energy-coupling factor transporter ATPase; |
899-1114 |
5.45e-19 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184200 [Multi-domain] Cd Length: 282 Bit Score: 89.47 E-value: 5.45e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTS---GTAYILGKDIRSE-MSSIRQN 974
Cdd:PRK13640 6 VEFKHVSFTYPDSKKPALNDISFSIPRGSWTALIGHNGSGKSTISKLINGLLLPDDnpnSKITVDGITLTAKtVWDIREK 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK13640 86 VGIVFQNpDNQFVGATVGDDVAFGLENRAVPRPEMIKIVRDVLADVGM-LDYIDSEPANLSGGQKQRVAIAGILAVEPKI 164
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGS 1114
Cdd:PRK13640 165 IILDESTSMLDPAGKEQILKLIrkLKKKNNLTVISITHDIDEAN-MADQVLVLDDGKLLAQGS 226
|
|
| ABCC_MRP_Like |
cd03228 |
ATP-binding cassette domain of multidrug resistance protein-like transporters; The MRP ... |
1914-2123 |
8.12e-19 |
|
ATP-binding cassette domain of multidrug resistance protein-like transporters; The MRP (Multidrug Resistance Protein)-like transporters are involved in drug, peptide, and lipid export. They belong to the subfamily C of the ATP-binding cassette (ABC) superfamily of transport proteins. The ABCC subfamily contains transporters with a diverse functional spectrum that includes ion transport, cell surface receptor, and toxin secretion activities. The MRP-like family, similar to all ABC proteins, have a common four-domain core structure constituted by two membrane-spanning domains, each composed of six transmembrane (TM) helices, and two nucleotide-binding domains (NBD). ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213195 [Multi-domain] Cd Length: 171 Bit Score: 85.90 E-value: 8.12e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:cd03228 1 IEFKNVSFSYPGRPKPVLKDVSLTIKPGEKVAIVGPSGSGKSTLLKLLLRLYDPTSGEILIDGVDLRDlDLESLRKNIAY 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQfdaitelltgreHVEFFAllrgvpekevGKVGEwairklglvkygekyasN-YSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:cd03228 81 VPQ------------DPFLFS----------GTIRE-----------------NiLSGGQRQRIAIARALLRDPPILILD 121
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2072 EPTTGMDPKARRFLWNcALSIVKEGRSVVLTSHSMEECEaLCTRMAIMVNGR 2123
Cdd:cd03228 122 EATSALDPETEALILE-ALRALAKGKTVIVIAHRLSTIR-DADRIIVLDDGR 171
|
|
| PRK09700 |
PRK09700 |
D-allose ABC transporter ATP-binding protein AlsA; |
899-1113 |
9.24e-19 |
|
D-allose ABC transporter ATP-binding protein AlsA;
Pssm-ID: 182036 [Multi-domain] Cd Length: 510 Bit Score: 92.15 E-value: 9.24e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLG-- 976
Cdd:PRK09700 6 ISMAGIGKSF--GPVHALKSVNLTVYPGEIHALLGENGAGKSTLMKVLSGIHEPTKGTITINNINYNKLDHKLAAQLGig 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFDMLTVEEHIwFYARLK-----GL-----SEKHVKAEMeqMALDVGLPPSkLKSKTSQLSGGMQRKLSVALA 1046
Cdd:PRK09700 84 IIYQELSVIDELTVLENL-YIGRHLtkkvcGVniidwREMRVRAAM--MLLRVGLKVD-LDEKVANLSISHKQMLEIAKT 159
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1047 FVGGSKVVILDEPTAGVdpySRRGIWELLLKYRQ----GRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:PRK09700 160 LMLDAKVIIMDEPTSSL---TNKEVDYLFLIMNQlrkeGTAIVYISHKLAEIRRICDRYTVMKDGSSVCSG 227
|
|
| ABCC_MsbA |
cd03251 |
ATP-binding cassette domain of the bacterial lipid flippase and related proteins, subfamily C; ... |
899-1108 |
9.63e-19 |
|
ATP-binding cassette domain of the bacterial lipid flippase and related proteins, subfamily C; MsbA is an essential ABC transporter, closely related to eukaryotic MDR proteins. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213218 [Multi-domain] Cd Length: 234 Bit Score: 87.67 E-value: 9.63e-19
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:cd03251 1 VEFKNVTFRYPGDGPPVLRDISLDIPAGETVALVGPSGSGKSTLVNLIPRFYDVDSGRILIDGHDVRDyTLASLRRQIGL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHIwFYARLkGLSEKHVKAEMEQ-------MALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGG 1050
Cdd:cd03251 81 VSQDVFLFND-TVAENI-AYGRP-GATREEVEEAARAanahefiMELPEGYD-TVIGERGVKLSGGQRQRIAIARALLKD 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1051 SKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTII-----LSThhMDEAdilgDRIAIISHGK 1108
Cdd:cd03251 157 PPILILDEATSALDTESERLVQAALERLMKNRTTFviahrLST--IENA----DRIVVLEDGK 213
|
|
| fecE |
PRK11231 |
Fe(3+) dicitrate ABC transporter ATP-binding protein FecE; |
901-1114 |
1.06e-18 |
|
Fe(3+) dicitrate ABC transporter ATP-binding protein FecE;
Pssm-ID: 183044 [Multi-domain] Cd Length: 255 Bit Score: 88.15 E-value: 1.06e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrsEMSSIRQ---NLGV 977
Cdd:PRK11231 5 TENLTVGY--GTKRILNDLSLSLPTGKITALIGPNGCGKSTLLKCFARLLTPQSGTVFLGDKPI--SMLSSRQlarRLAL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEE--------HIWFYARLKGLSEKHVKAEMEQMALDvglppsKLKSK-TSQLSGGMQRKLSVALAFV 1048
Cdd:PRK11231 81 LPQHHLTPEGITVRElvaygrspWLSLWGRLSAEDNARVNQAMEQTRIN------HLADRrLTDLSGGQRQRAFLAMVLA 154
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK11231 155 QDTPVVLLDEPTTYLDINHQVELMRLMRELNTqGKTVVTVLHDLNQASRYCDHLVVLANGHVMAQGT 221
|
|
| cbiO |
PRK13635 |
energy-coupling factor ABC transporter ATP-binding protein; |
899-1109 |
1.25e-18 |
|
energy-coupling factor ABC transporter ATP-binding protein;
Pssm-ID: 184195 [Multi-domain] Cd Length: 279 Bit Score: 88.53 E-value: 1.25e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK--------DIRSEMSS 970
Cdd:PRK13635 6 IRVEHISFRYPDAATYALKDVSFSVYEGEWVAIVGHNGSGKSTLAKLLNGLLLPEAGTITVGGMvlseetvwDVRRQVGM 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQNlgvcPQHNvlFDMLTVEEHIWFyarlkGLSEKHVkaEMEQM------ALD-VGLpPSKLKSKTSQLSGGmqRKLSV 1043
Cdd:PRK13635 86 VFQN----PDNQ--FVGATVQDDVAF-----GLENIGV--PREEMvervdqALRqVGM-EDFLNREPHRLSGG--QKQRV 149
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1044 ALAFVGGS--KVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:PRK13635 150 AIAGVLALqpDIIILDEATSMLDPRGRREVLETVrqLKEQKGITVLSITHDLDEA-AQADRVIVMNKGEI 218
|
|
| PRK14246 |
PRK14246 |
phosphate ABC transporter ATP-binding protein; Provisional |
928-1128 |
1.37e-18 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172734 [Multi-domain] Cd Length: 257 Bit Score: 87.80 E-value: 1.37e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 928 ITSFLGHNGAGKTTTMSILTGLFP------PTSGTAYILGKDI-RSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWFYARL 1000
Cdd:PRK14246 38 IFGIMGPSGSGKSTLLKVLNRLIEiydskiKVDGKVLYFGKDIfQIDAIKLRKEVGMVFQQPNPFPHLSIYDNIAYPLKS 117
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 KGLSEK-HVKAEMEQMALDVGLPPS---KLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLL 1076
Cdd:PRK14246 118 HGIKEKrEIKKIVEECLRKVGLWKEvydRLNSPASQLSGGQQQRLTIARALALKPKVLLMDEPTSMIDIVNSQAIEKLIT 197
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1077 KYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS--LFL--KNQLGTGYYL 1128
Cdd:PRK14246 198 ELKNEIAIVIVSHNPQQVARVADYVAFLYNGELVEWGSSneIFTspKNELTEKYVI 253
|
|
| 40850658_otr |
NF000106 |
oxytetracycline efflux ABC transporter Otr(C) ATP-binding subunit; |
896-1123 |
1.39e-18 |
|
oxytetracycline efflux ABC transporter Otr(C) ATP-binding subunit;
Pssm-ID: 411078 [Multi-domain] Cd Length: 351 Bit Score: 89.79 E-value: 1.39e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 896 RLGVSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAgktttmSILTGLFPptsgtAYILGKDIR---------- 965
Cdd:NF000106 11 RNAVEVRGLVKHF--GEVKAVDGVDLDVREGTVLGVLGP*GA------A**RGALP-----AHV*GPDAGrrpwrf*twc 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 966 SEMSSIRQNLGVC-PQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSkTSQLSGGMQRKLSVA 1044
Cdd:NF000106 78 ANRRALRRTIG*HrPVR*GRRESFSGRENLYMIGR*LDLSRKDARARADELLERFSLTEAAGRA-AAKYSGGMRRRLDLA 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRRGIW-ELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKNQLG 1123
Cdd:NF000106 157 ASMIGRPAVLYLDEPTTGLDPRTRNEVWdEVRSMVRDGATVLLTTQYMEEAEQLAHELTVIDRGRVIADGKVDELKTKVG 236
|
|
| cbiO |
PRK13652 |
cobalt transporter ATP-binding subunit; Provisional |
1913-2133 |
1.56e-18 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 172200 [Multi-domain] Cd Length: 277 Bit Score: 87.94 E-value: 1.56e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL-SNIHEVHQNMG 1991
Cdd:PRK13652 3 LIETRDLCYSYSGSKE-ALNNINFIAPRNSRIAVIGPNGAGKSTLFRHFNGILKPTSGSVLIRGEPITkENIREVRKFVG 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQF--DAITELlTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:PRK13652 82 LVFQNpdDQIFSP-TVEQDIAFGPINLGLDEETVAHRVSSALHMLGLEELRDRVPHHLSGGEKKRVAIAGVIAMEPQVLV 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2070 LDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK13652 161 LDEPTAGLDPQGVKELIDFLNDLPETyGMTVIFSTHQLDLVPEMADYIYVMDKGRIVAYGTVEEI 225
|
|
| tauB |
PRK11248 |
taurine ABC transporter ATP-binding subunit; |
913-1107 |
1.66e-18 |
|
taurine ABC transporter ATP-binding subunit;
Pssm-ID: 183056 [Multi-domain] Cd Length: 255 Bit Score: 87.45 E-value: 1.66e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 913 KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR---SEMSSIRQNLGVCPQHNVLfdmlt 989
Cdd:PRK11248 14 KPALEDINLTLESGELLVVLGPSGCGKTTLLNLIAGFVPYQHGSITLDGKPVEgpgAERGVVFQNEGLLPWRNVQ----- 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 990 veEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRR 1069
Cdd:PRK11248 89 --DNVAFGLQLAGVEKMQRLEIAHQMLKKVGLEGAE-KRYIWQLSGGQRQRVGIARALAANPQLLLLDEPFGALDAFTRE 165
|
170 180 190 200
....*....|....*....|....*....|....*....|
gi 568925948 1070 GIWELLLK--YRQGRTIILSTHHMDEADILGDRIAIISHG 1107
Cdd:PRK11248 166 QMQTLLLKlwQETGKQVLLITHDIEEAVFMATELVLLSPG 205
|
|
| nickel_nikE |
TIGR02769 |
nickel import ATP-binding protein NikE; This family represents the NikE subunit of a ... |
899-1109 |
1.83e-18 |
|
nickel import ATP-binding protein NikE; This family represents the NikE subunit of a multisubunit nickel import ABC transporter complex. Nickel, once imported, may be used in urease and in certain classes of hydrogenase and superoxide dismutase. [Transport and binding proteins, Cations and iron carrying compounds]
Pssm-ID: 131816 [Multi-domain] Cd Length: 265 Bit Score: 87.55 E-value: 1.83e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGM-------KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSE 967
Cdd:TIGR02769 3 LEVRDVTHTYRTGGlfgakqrAPVLTNVSLSIEEGETVGLLGRSGCGKSTLARLLLGLEKPAQGTVSFRGQDLyqldRKQ 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MSSIRQNLGVCPQ--HNVLFDMLTVEEHIWFYAR-LKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVA 1044
Cdd:TIGR02769 83 RRAFRRDVQLVFQdsPSAVNPRMTVRQIIGEPLRhLTSLDESEQKARIAELLDMVGLRSEDADKLPRQLSGGQLQRINIA 162
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:TIGR02769 163 RALAVKPKLIVLDEAVSNLDMVLQAVILELLRKLQQafGTAYLFITHDLRLVQSFCQRVAVMDKGQI 229
|
|
| COG4674 |
COG4674 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
900-1114 |
1.97e-18 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 443710 [Multi-domain] Cd Length: 250 Bit Score: 87.09 E-value: 1.97e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC- 978
Cdd:COG4674 12 YVEDLTVSF-DGFK-ALNDLSLYVDPGELRVIIGPNGAGKTTLMDVITGKTRPDSGSVLFGGTDLTGLDEHEIARLGIGr 89
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 ----PqhNVlFDMLTVEEH----------IW--FYARLKGlsekHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLS 1042
Cdd:COG4674 90 kfqkP--TV-FEELTVFENlelalkgdrgVFasLFARLTA----EERDRIEEVLETIGL-TDKADRLAGLLSHGQKQWLE 161
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1043 VALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:COG4674 162 IGMLLAQDPKLLLLDEPVAGMTDAETERTAELLKSLAGKHSVVVVEHDMEFVRQIARKVTVLHQGSVLAEGS 233
|
|
| cbiO |
PRK13650 |
energy-coupling factor transporter ATPase; |
899-1109 |
2.03e-18 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184209 [Multi-domain] Cd Length: 279 Bit Score: 87.86 E-value: 2.03e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNL-VKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLG 976
Cdd:PRK13650 5 IEVKNLtFKYKEDQEKYTLNDVSFHVKQGEWLSIIGHNGSGKSTTVRLIDGLLEAESGQIIIDGDLLTEEnVWDIRHKIG 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQmALD-VGLPPSKLKsKTSQLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:PRK13650 85 MVFQNpDNQFVGATVEDDVAFGLENKGIPHEEMKERVNE-ALElVGMQDFKER-EPARLSGGQKQRVAIAGAVAMRPKII 162
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:PRK13650 163 ILDEATSMLDPEGRLELIKTIKGIRDdyQMTVISITHDLDEV-ALSDRVLVMKNGQV 218
|
|
| cbiO |
PRK13637 |
energy-coupling factor transporter ATPase; |
1914-2130 |
2.27e-18 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237455 [Multi-domain] Cd Length: 287 Bit Score: 87.80 E-value: 2.27e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRK---RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS---NIHEVH 1987
Cdd:PRK13637 3 IKIENLTHIYMEGtpfEKKALDNVNIEIEDGEFVGLIGHTGSGKSTLIQHLNGLLKPTSGKIIIDGVDITDkkvKLSDIR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNMG----YcPQFDAITEllTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLV--KYGEKYASNYSGGNKRKLSTAMAL 2061
Cdd:PRK13637 83 KKVGlvfqY-PEYQLFEE--TIEKDIAFGPINLGLSEEEIENRVKRAMNIVGLDyeDYKDKSPFELSGGQKRRVAIAGVV 159
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2062 IGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSV 2130
Cdd:PRK13637 160 AMEPKILILDEPTAGLDPKGRDEILNKIKELHKEyNMTIILVSHSMEDVAKLADRIIVMNKGKCELQGTP 229
|
|
| FtsE |
COG2884 |
Cell division ATPase FtsE [Cell cycle control, cell division, chromosome partitioning]; |
1913-2107 |
2.83e-18 |
|
Cell division ATPase FtsE [Cell cycle control, cell division, chromosome partitioning];
Pssm-ID: 442130 [Multi-domain] Cd Length: 223 Bit Score: 85.87 E-value: 2.83e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkPAVDRICIGIPPGE-CFgLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI--LSN--IHEVH 1987
Cdd:COG2884 1 MIRFENVSKRYPGGR-EALSDVSLEIEKGEfVF-LTGPSGAGKSTLLKLLYGEERPTSGQVLVNGQDLsrLKRreIPYLR 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNMGYCPQ-FdaitELLTGR---EHVEFFALLRGVPEKEVGK-VGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALI 2062
Cdd:COG2884 79 RRIGVVFQdF----RLLPDRtvyENVALPLRVTGKSRKEIRRrVRE-VLDLVGLSDKAKALPHELSGGEQQRVAIARALV 153
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 2063 GGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSME 2107
Cdd:COG2884 154 NRPELLLADEPTGNLDPETSWEIMELLEEINRRGTTVLIATHDLE 198
|
|
| PRK10908 |
PRK10908 |
cell division ATP-binding protein FtsE; |
899-1109 |
2.84e-18 |
|
cell division ATP-binding protein FtsE;
Pssm-ID: 182829 [Multi-domain] Cd Length: 222 Bit Score: 86.08 E-value: 2.84e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQN 974
Cdd:PRK10908 2 IRFEHVSKAYLGG-RQALQGVTFHMRPGEMAFLTGHSGAGKSTLLKLICGIERPSAGKIWFSGHDItrlkNREVPFLRRQ 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQ-HNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQmALD-VGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:PRK10908 81 IGMIFQdHHLLMDR-TVYDNVAIPLIIAGASGDDIRRRVSA-ALDkVGLL-DKAKNFPIQLSGGEQQRVGIARAVVNKPA 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK10908 158 VLLADEPTGNLDDALSEGILRLFEEFnRVGVTVLMATHDIGLISRRSYRMLTLSDGHL 215
|
|
| PRK10535 |
PRK10535 |
macrolide ABC transporter ATP-binding protein/permease MacB; |
901-1109 |
3.41e-18 |
|
macrolide ABC transporter ATP-binding protein/permease MacB;
Pssm-ID: 182528 [Multi-domain] Cd Length: 648 Bit Score: 91.32 E-value: 3.41e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYRDG-MKVAV-DGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSS------IR 972
Cdd:PRK10535 7 LKDIRRSYPSGeEQVEVlKGISLDIYAGEMVAIVGASGSGKSTLMNILGCLDKPTSGTYRVAGQDV-ATLDAdalaqlRR 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:PRK10535 86 EHFGFIFQRYHLLSHLTAAQNVEVPAVYAGLERKQRLLRAQELLQRLGLE-DRVEYQPSQLSGGQQQRVSIARALMNGGQ 164
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADiLGDRIAIISHGKL 1109
Cdd:PRK10535 165 VILADEPTGALDSHSGEEVMAILHQLRdRGHTVIIVTHDPQVAA-QAERVIEIRDGEI 221
|
|
| ABC_ModC_molybdenum_transporter |
cd03297 |
ATP-binding cassette domain of the molybdenum transport system; ModC is an ABC-type ... |
925-1109 |
4.28e-18 |
|
ATP-binding cassette domain of the molybdenum transport system; ModC is an ABC-type transporter and the ATPase component of a molybdate transport system that also includes the periplasmic binding protein ModA and the membrane protein ModB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213264 [Multi-domain] Cd Length: 214 Bit Score: 85.04 E-value: 4.28e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 925 EGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK-----DIRSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWFYAR 999
Cdd:cd03297 22 NEEVTGIFGASGAGKSTLLRCIAGLEKPDGGTIVLNGTvlfdsRKKINLPPQQRKIGLVFQQYALFPHLNVRENLAFGLK 101
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1000 LKGLSEKHVKAE--MEQMALDvglppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL-- 1075
Cdd:cd03297 102 RKRNREDRISVDelLDLLGLD-----HLLNRYPAQLSGGEKQRVALARALAAQPELLLLDEPFSALDRALRLQLLPELkq 176
|
170 180 190
....*....|....*....|....*....|....
gi 568925948 1076 LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:cd03297 177 IKKNLNIPVIFVTHDLSEAEYLADRIVVMEDGRL 210
|
|
| MsbA_lipidA |
TIGR02203 |
lipid A export permease/ATP-binding protein MsbA; This family consists of a single polypeptide ... |
899-1109 |
4.32e-18 |
|
lipid A export permease/ATP-binding protein MsbA; This family consists of a single polypeptide chain transporter in the ATP-binding cassette (ABC) transporter family, MsbA, which exports lipid A. It may also act in multidrug resistance. Lipid A, a part of lipopolysaccharide, is found in the outer leaflet of the outer membrane of most Gram-negative bacteria. Members of this family are restricted to the Proteobacteria (although lipid A is more broadly distributed) and often are clustered with lipid A biosynthesis genes. [Cell envelope, Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides, Transport and binding proteins, Other]
Pssm-ID: 131258 [Multi-domain] Cd Length: 571 Bit Score: 90.55 E-value: 4.32e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:TIGR02203 331 VEFRNVTFRYPGRDRPALDSISLVIEPGETVALVGRSGSGKSTLVNLIPRFYEPDSGQILLDGHDLADyTLASLRRQVAL 410
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHIwFYARLKGLSEKHVKAEMEQM-ALDV--GLP---PSKLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:TIGR02203 411 VSQDVVLFND-TIANNI-AYGRTEQADRAEIERALAAAyAQDFvdKLPlglDTPIGENGVLLSGGQRQRLAIARALLKDA 488
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMdeADILG-DRIAIISHGKL 1109
Cdd:TIGR02203 489 PILILDEATSALDNESERLVQAALERLMQGRTTLVIAHRL--STIEKaDRIVVMDDGRI 545
|
|
| ABC_Carb_Monos_I |
cd03216 |
First domain of the ATP-binding cassette component of monosaccharide transport system; This ... |
1914-2123 |
5.80e-18 |
|
First domain of the ATP-binding cassette component of monosaccharide transport system; This family represents the domain I of the carbohydrate uptake proteins that transport only monosaccharides (Monos). The Carb_Monos family is involved in the uptake of monosaccharides, such as pentoses (such as xylose, arabinose, and ribose) and hexoses (such as xylose, arabinose, and ribose), that cannot be broken down to simple sugars by hydrolysis. Pentoses include xylose, arabinose, and ribose. Important hexoses include glucose, galactose, and fructose. In members of the Carb_monos family, the single hydrophobic gene product forms a homodimer while the ABC protein represents a fusion of two nucleotide-binding domains. However, it is assumed that two copies of the ABC domains are present in the assembled transporter.
Pssm-ID: 213183 [Multi-domain] Cd Length: 163 Bit Score: 83.25 E-value: 5.80e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknSILSNIHEVHqnmgyc 1993
Cdd:cd03216 1 LELRGITKRFGGVK--ALDGVSLSVRRGEVHALLGENGAGKSTLMKILSGLYKPDSG-------EILVDGKEVS------ 65
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 pqFDAITElltgrehveffALLRGVpekevgkvgewairklGLVkygekyaSNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03216 66 --FASPRD-----------ARRAGI----------------AMV-------YQLSVGERQMVEIARALARNARLLILDEP 109
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03216 110 TAALTPAEVERLFKVIRRLRAQGVAVIFISHRLDEVFEIADRVTVLRDGR 159
|
|
| ABC_CcmA_heme_exporter |
cd03231 |
Cytochrome c biogenesis ATP-binding export protein; CcmA, the ATP-binding component of the ... |
909-1090 |
5.82e-18 |
|
Cytochrome c biogenesis ATP-binding export protein; CcmA, the ATP-binding component of the bacterial CcmAB transporter. The CCM family is involved in bacterial cytochrome c biogenesis. Cytochrome c maturation in E. coli requires the ccm operon, which encodes eight membrane proteins (CcmABCDEFGH). CcmE is a periplasmic heme chaperon that binds heme covalently and transfers it onto apocytochrome c in the presence of CcmF, CcmG, and CcmH. The CcmAB proteins represent an ABC transporter and the CcmCD proteins participate in heme transfer to CcmE.
Pssm-ID: 213198 [Multi-domain] Cd Length: 201 Bit Score: 84.47 E-value: 5.82e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDML 988
Cdd:cd03231 10 RDG-RALFSGLSFTLAAGEALQVTGPNGSGKTTLLRILAGLSPPLAGRVLLNGGPLDFQRDSIARGLLYLGHAPGIKTTL 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWFYARLkglsekHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSR 1068
Cdd:cd03231 89 SVLENLRFWHAD------HSDEQVEEALARVGLNGFE-DRPVAQLSAGQQRRVALARLLLSGRPLWILDEPTTALDKAGV 161
|
170 180
....*....|....*....|...
gi 568925948 1069 RGIWELLLKY-RQGRTIILSTHH 1090
Cdd:cd03231 162 ARFAEAMAGHcARGGMVVLTTHQ 184
|
|
| LivF |
COG0410 |
ABC-type branched-chain amino acid transport system, ATPase component LivF [Amino acid ... |
1913-2123 |
6.71e-18 |
|
ABC-type branched-chain amino acid transport system, ATPase component LivF [Amino acid transport and metabolism];
Pssm-ID: 440179 [Multi-domain] Cd Length: 236 Bit Score: 85.03 E-value: 6.71e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHE-VHQNM 1990
Cdd:COG0410 3 MLEVENLHAGY--GGIHVLHGVSLEVEEGEIVALLGRNGAGKTTLLKAISGLLPPRSGSIRFDGEDITGlPPHRiARLGI 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAITELLTGREHVEFFALLRGVPEKevgkvGEWAIRKLG-----LVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:COG0410 81 GYVPEGRRIFPSLTVEENLLLGAYARRDRAE-----VRADLERVYelfprLKERRRQRAGTLSGGEQQMLAIGRALMSRP 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG0410 156 KLLLLDEPSLGLAPLIVEEIFEIIRRLNREGVTILLVEQNARFALEIADRAYVLERGR 213
|
|
| cbiO |
PRK13631 |
cobalt transporter ATP-binding subunit; Provisional |
899-1114 |
7.41e-18 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237451 [Multi-domain] Cd Length: 320 Bit Score: 87.21 E-value: 7.41e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMK---VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYI----LGKDIRSEMSSI 971
Cdd:PRK13631 22 LRVKNLYCVFDEKQEnelVALNNISYTFEKNKIYFIIGNSGSGKSTLVTHFNGLIKSKYGTIQVgdiyIGDKKNNHELIT 101
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 972 -------------RQNLGVC---PQHNVLFDmlTVEEHIWFYARLKGLS--EKHVKAE--MEQMaldvGLPPSKLKSKTS 1031
Cdd:PRK13631 102 npyskkiknfkelRRRVSMVfqfPEYQLFKD--TIEKDIMFGPVALGVKksEAKKLAKfyLNKM----GLDDSYLERSPF 175
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1032 QLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKLC 1110
Cdd:PRK13631 176 GLSGGQKRRVAIAGILAIQPEILIFDEPTAGLDPKGEHEMMQLILDAKaNNKTVFVITHTMEHVLEVADEVIVMDKGKIL 255
|
....
gi 568925948 1111 CVGS 1114
Cdd:PRK13631 256 KTGT 259
|
|
| ccmA |
TIGR01189 |
heme ABC exporter, ATP-binding protein CcmA; This model describes the cyt c biogenesis protein ... |
1914-2104 |
7.78e-18 |
|
heme ABC exporter, ATP-binding protein CcmA; This model describes the cyt c biogenesis protein encoded by ccmA in bacteria. An exception is, an arabidopsis protein. Quite likely this is encoded by an organelle. Bacterial c-type cytocromes are located on the periplasmic side of the cytoplasmic membrane. Several gene products encoded in a locus designated as 'ccm' are implicated in the transport and assembly of the functional cytochrome C. This cluster includes genes: ccmA;B;C;D;E;F;G and H. The posttranslational pathway includes the transport of heme moiety, the secretion of the apoprotein and the covalent attachment of the heme with the apoprotein. The proteins ccmA and B represent an ABC transporter; ccmC and D participate in heme transfer to ccmE, which function as a periplasmic heme chaperone. The presence of ccmF, G and H is suggested to be obligatory for the final functional assembly of cytochrome c. [Protein fate, Protein and peptide secretion and trafficking, Transport and binding proteins, Other]
Pssm-ID: 273491 [Multi-domain] Cd Length: 198 Bit Score: 83.95 E-value: 7.78e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:TIGR01189 1 LAARNLA--CSRGERMLFEGLSFTLNAGEALQVTGPNGIGKTTLLRILAGLLRPDSGEVRWNGTPLAEQRDEPHENILYL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVgkvgEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:TIGR01189 79 GHLPGLKPELSALENLHFWAAIHGGAQRTI----EDALAAVGLTGFEDLPAAQLSAGQQRRLALARLWLSRRPLWILDEP 154
|
170 180 190
....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:TIGR01189 155 TTALDKAGVALLAGLLRAHLARGGIVLLTTH 185
|
|
| SunT |
COG2274 |
ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase ... |
1887-2144 |
8.02e-18 |
|
ABC-type bacteriocin/lantibiotic exporters, contain an N-terminal double-glycine peptidase domain [Defense mechanisms];
Pssm-ID: 441875 [Multi-domain] Cd Length: 711 Bit Score: 90.28 E-value: 8.02e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1887 LPPlnDEDEDVRRERQRILDGGgqndiLEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP 1966
Cdd:COG2274 454 LPP--EREEGRSKLSLPRLKGD-----IELENVSFRYPGDSPPVLDNISLTIKPGERVAIVGRSGSGKSTLLKLLLGLYE 526
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1967 VTRGDAFLNKNSILS-NIHEVHQNMGYCPQfDaiTELLTG--REHVEFFAllRGVPEKEVgkvgEWAIRKLGLVKY---- 2039
Cdd:COG2274 527 PTSGRILIDGIDLRQiDPASLRRQIGVVLQ-D--VFLFSGtiRENITLGD--PDATDEEI----IEAARLAGLHDFieal 597
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2040 --------GEKyASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSHSMeECEA 2111
Cdd:COG2274 598 pmgydtvvGEG-GSNLSGGQRQRLAIARALLRNPRILILDEATSALDAETEAIILE-NLRRLLKGRTVIIIAHRL-STIR 674
|
250 260 270
....*....|....*....|....*....|...
gi 568925948 2112 LCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIV 2144
Cdd:COG2274 675 LADRIIVLDKGRIVEDGTHEELLARKGLYAELV 707
|
|
| ABCC_TAP |
cd03248 |
ATP-binding cassette domain of the Transporter Associated with Antigen Processing, subfamily C; ... |
892-1109 |
9.74e-18 |
|
ATP-binding cassette domain of the Transporter Associated with Antigen Processing, subfamily C; TAP (Transporter Associated with Antigen Processing) is essential for peptide delivery from the cytosol into the lumen of the endoplasmic reticulum (ER), where these peptides are loaded on major histocompatibility complex (MHC) I molecules. Loaded MHC I leave the ER and display their antigenic cargo on the cell surface to cytotoxic T cells. Subsequently, virus-infected or malignantly transformed cells can be eliminated. TAP belongs to the large family of ATP-binding cassette (ABC) transporters, which translocate a vast variety of solutes across membranes.
Pssm-ID: 213215 [Multi-domain] Cd Length: 226 Bit Score: 84.44 E-value: 9.74e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 892 PTHLRLGVSIQNLVKVYRDGMKVAV-DGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMS 969
Cdd:cd03248 5 PDHLKGIVKFQNVTFAYPTRPDTLVlQDVSFTLHPGEVTALVGPSGSGKSTVVALLENFYQPQGGQVLLDGKPISQyEHK 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 970 SIRQNLGVCPQHNVLFDMlTVEEHIWFyaRLKGLSEKHVKA------------EMEQ-MALDVGlppsklkSKTSQLSGG 1036
Cdd:cd03248 85 YLHSKVSLVGQEPVLFAR-SLQDNIAY--GLQSCSFECVKEaaqkahahsfisELASgYDTEVG-------EKGSQLSGG 154
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1037 MQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKL 1109
Cdd:cd03248 155 QKQRVAIARALIRNPQVLILDEATSALDAESEQQVQQALYDWPERRTVLVIAHRLSTVE-RADQILVLDGGRI 226
|
|
| TagH |
COG1134 |
ABC-type polysaccharide/polyol phosphate transport system, ATPase component [Carbohydrate ... |
899-1114 |
1.01e-17 |
|
ABC-type polysaccharide/polyol phosphate transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440749 [Multi-domain] Cd Length: 245 Bit Score: 84.75 E-value: 1.01e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYR------DGMK--------------VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAY 958
Cdd:COG1134 5 IEVENVSKSYRlyhepsRSLKelllrrrrtrreefWALKDVSFEVERGESVGIIGRNGAGKSTLLKLIAGILEPTSGRVE 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 959 ILGKdirseMSSIrqnLGVcpqhNVLFDM-LTVEEHIWFYARLKGLSEKHVK---------AEMEQmALDvgLPpskLKS 1028
Cdd:COG1134 85 VNGR-----VSAL---LEL----GAGFHPeLTGRENIYLNGRLLGLSRKEIDekfdeivefAELGD-FID--QP---VKT 146
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1029 ktsqLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPY----SRRGIWELLlkyRQGRTIILSTHHMDEADILGDRIAII 1104
Cdd:COG1134 147 ----YSSGMRARLAFAVATAVDPDILLVDEVLAVGDAAfqkkCLARIRELR---ESGRTVIFVSHSMGAVRRLCDRAIWL 219
|
250
....*....|
gi 568925948 1105 SHGKLCCVGS 1114
Cdd:COG1134 220 EKGRLVMDGD 229
|
|
| fbpC |
PRK11432 |
ferric ABC transporter ATP-binding protein; |
899-1123 |
1.15e-17 |
|
ferric ABC transporter ATP-binding protein;
Pssm-ID: 183133 [Multi-domain] Cd Length: 351 Bit Score: 87.08 E-value: 1.15e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKvyRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSemSSIrQNLGVC 978
Cdd:PRK11432 7 VVLKNITK--RFGSNTVIDNLNLTIKQGTMVTLLGPSGCGKTTVLRLVAGLEKPTEGQIFIDGEDVTH--RSI-QQRDIC 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 P--QHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEM-EQMALdVGLppSKLKSK-TSQLSGGMQRKLSVALAFVGGSKVV 1054
Cdd:PRK11432 82 MvfQSYALFPHMSLGENVGYGLKMLGVPKEERKQRVkEALEL-VDL--AGFEDRyVDQISGGQQQRVALARALILKPKVL 158
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1055 ILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS---------SLFLKNQLG 1123
Cdd:PRK11432 159 LFDEPLSNLDANLRRSMREKIreLQQQFNITSLYVTHDQSEAFAVSDTVIVMNKGKIMQIGSpqelyrqpaSRFMASFMG 238
|
|
| PRK10584 |
PRK10584 |
putative ABC transporter ATP-binding protein YbbA; Provisional |
918-1089 |
1.31e-17 |
|
putative ABC transporter ATP-binding protein YbbA; Provisional
Pssm-ID: 182569 [Multi-domain] Cd Length: 228 Bit Score: 84.06 E-value: 1.31e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 918 GLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-------RSEMSSirQNLGVCPQHNVLFDMLTV 990
Cdd:PRK10584 28 GVELVVKRGETIALIGESGSGKSTLLAILAGLDDGSSGEVSLVGQPLhqmdeeaRAKLRA--KHVGFVFQSFMLIPTLNA 105
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 991 EEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRG 1070
Cdd:PRK10584 106 LENVELPALLRGESSRQSRNGAKALLEQLGLG-KRLDHLPAQLSGGEQQRVALARAFNGRPDVLFADEPTGNLDRQTGDK 184
|
170 180
....*....|....*....|.
gi 568925948 1071 IWELL--LKYRQGRTIILSTH 1089
Cdd:PRK10584 185 IADLLfsLNREHGTTLILVTH 205
|
|
| glnQ |
PRK09493 |
glutamine ABC transporter ATP-binding protein GlnQ; |
899-1109 |
1.53e-17 |
|
glutamine ABC transporter ATP-binding protein GlnQ;
Pssm-ID: 181906 [Multi-domain] Cd Length: 240 Bit Score: 84.37 E-value: 1.53e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS---IRQNL 975
Cdd:PRK09493 2 IEFKNVSKHF--GPTQVLHNIDLNIDQGEVVVIIGPSGSGKSTLLRCINKLEEITSGDLIVDGLKVNDPKVDerlIRQEA 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQHNVLFDMLTVEEHIWFYA-RLKGLSekhvKAEMEQMALD----VGLPpSKLKSKTSQLSGGMQRKLSVALAFVGG 1050
Cdd:PRK09493 80 GMVFQQFYLFPHLTALENVMFGPlRVRGAS----KEEAEKQAREllakVGLA-ERAHHYPSELSGGQQQRVAIARALAVK 154
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1051 SKVVILDEPTAGVDPYSRRgiwELLLKYR----QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK09493 155 PKLMLFDEPTSALDPELRH---EVLKVMQdlaeEGMTMVIVTHEIGFAEKVASRLIFIDKGRI 214
|
|
| potG |
PRK11607 |
putrescine ABC transporter ATP-binding subunit PotG; |
1913-2183 |
1.60e-17 |
|
putrescine ABC transporter ATP-binding subunit PotG;
Pssm-ID: 183226 [Multi-domain] Cd Length: 377 Bit Score: 87.20 E-value: 1.60e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSiLSNIHEVHQNMGY 1992
Cdd:PRK11607 19 LLEIRNLTKSF--DGQHAVDDVSLTIYKGEIFALLGASGCGKSTLLRMLAGFEQPTAGQIMLDGVD-LSHVPPYQRPINM 95
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITELLTGREHVEFFALLRGVPEKEV-GKVGEwairKLGLV---KYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:PRK11607 96 MFQSYALFPHMTVEQNIAFGLKQDKLPKAEIaSRVNE----MLGLVhmqEFAKRKPHQLSGGQRQRVALARSLAKRPKLL 171
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKnrfgdgytivvri 2147
Cdd:PRK11607 172 LLDEPMGALDKKLRDRMQLEVVDILERvGVTCVMVTHDQEEAMTMAGRIAIMNRGKFVQIGEPEEIY------------- 238
|
250 260 270
....*....|....*....|....*....|....*...
gi 568925948 2148 agSNPDLKPVQEFFGLA--FPGsVLKEKHRNMLQYQLP 2183
Cdd:PRK11607 239 --EHPTTRYSAEFIGSVnvFEG-VLKERQEDGLVIDSP 273
|
|
| bacteriocin_ABC |
TIGR01193 |
ABC-type bacteriocin transporter; This model describes ABC-type bacteriocin transporter. The ... |
915-1131 |
1.63e-17 |
|
ABC-type bacteriocin transporter; This model describes ABC-type bacteriocin transporter. The amino terminal domain (pfam03412) processes the N-terminal leader peptide from the bacteriocin while C-terminal domains resemble ABC transporter membrane protein and ATP-binding cassette domain. In general, bacteriocins are agents which are responsible for killing or inhibiting the closely related species or even different strains of the same species. Bacteriocins are usually encoded by bacterial plasmids. Bacteriocins are named after the species and hence in literature one encounters various names e.g., leucocin from Leuconostic geldium; pedicocin from Pedicoccus acidilactici; sakacin from Lactobacillus sake etc. [Protein fate, Protein and peptide secretion and trafficking, Protein fate, Protein modification and repair, Transport and binding proteins, Other]
Pssm-ID: 130261 [Multi-domain] Cd Length: 708 Bit Score: 89.41 E-value: 1.63e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMlTVEEH 993
Cdd:TIGR01193 489 ILSDISLTIKMNSKTTIVGMSGSGKSTLAKLLVGFFQARSGEILLNGFSLKDiDRHTLRQFINYLPQEPYIFSG-SILEN 567
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 994 IWFYAR--------LKGLSEKHVKAEMEQMALDVGlppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:TIGR01193 568 LLLGAKenvsqdeiWAACEIAEIKDDIENMPLGYQ---TELSEEGSSISGGQKQRIALARALLTDSKVLILDESTSNLDT 644
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1066 YSRRGIWELLLKYrQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGSSLFLKNQlgTGYYLTLV 1131
Cdd:TIGR01193 645 ITEKKIVNNLLNL-QDKTIIFVAHRLSVAK-QSDKIIVLDHGKIIEQGSHDELLDR--NGFYASLI 706
|
|
| cbiO |
PRK13642 |
energy-coupling factor transporter ATPase; |
899-1109 |
2.86e-17 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184202 [Multi-domain] Cd Length: 277 Bit Score: 84.37 E-value: 2.86e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKV-AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLG 976
Cdd:PRK13642 5 LEVENLVFKYEKESDVnQLNGVSFSITKGEWVSIIGQNGSGKSTTARLIDGLFEEFEGKVKIDGELLTAEnVWNLRRKIG 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKsKTSQLSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:PRK13642 85 MVFQNpDNQFVGATVEDDVAFGMENQGIPREEMIKRVDEALLAVNMLDFKTR-EPARLSGGQKQRVAVAGIIALRPEIII 163
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1056 LDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:PRK13642 164 LDESTSMLDPTGRQEIMRVIheIKEKYQLTVLSITHDLDEA-ASSDRILVMKAGEI 218
|
|
| cbiO |
PRK13639 |
cobalt transporter ATP-binding subunit; Provisional |
1913-2123 |
2.94e-17 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184199 [Multi-domain] Cd Length: 275 Bit Score: 84.36 E-value: 2.94e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI---LSNIHEVHQN 1989
Cdd:PRK13639 1 ILETRDLKYSYP-DGTEALKGINFKAEKGEMVALLGPNGAGKSTLFLHFNGILKPTSGEVLIKGEPIkydKKSLLEVRKT 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQfDAITELL--TGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:PRK13639 80 VGIVFQ-NPDDQLFapTVEEDVAFGPLNLGLSKEEVEKRVKEALKAVGMEGFENKPPHHLSGGQKKRVAIAGILAMKPEI 158
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13639 159 IVLDEPTSGLDPMGASQIMKLLYDLNKEGITIIISTHDVDLVPVYADKVYVMSDGK 214
|
|
| PRK10895 |
PRK10895 |
lipopolysaccharide ABC transporter ATP-binding protein; Provisional |
1913-2129 |
2.95e-17 |
|
lipopolysaccharide ABC transporter ATP-binding protein; Provisional
Pssm-ID: 182817 [Multi-domain] Cd Length: 241 Bit Score: 83.40 E-value: 2.95e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHE-VHQNM 1990
Cdd:PRK10895 3 TLTAKNLAKAYKGRR--VVEDVSLTVNSGEIVGLLGPNGAGKTTTFYMVVGIVPRDAGNIIIDDEDIsLLPLHArARRGI 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKV-GEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:PRK10895 81 GYLPQEASIFRRLSVYDNLMAVLQIRDDLSAEQREDrANELMEEFHIEHLRDSMGQSLSGGERRRVEIARALAANPKFIL 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2070 LDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGS 2129
Cdd:PRK10895 161 LDEPFAGVDPISVIDIKRIIEHLRDSGLGVLITDHNVRETLAVCERAYIVSQGHLIAHGT 220
|
|
| hmuV |
PRK13548 |
hemin importer ATP-binding subunit; Provisional |
1914-2131 |
3.29e-17 |
|
hemin importer ATP-binding subunit; Provisional
Pssm-ID: 237422 [Multi-domain] Cd Length: 258 Bit Score: 83.67 E-value: 3.29e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:PRK13548 3 LEARNLS--VRLGGRTLLDDVSLTLRPGEVVAILGPNGAGKSTLLRALSGELSPDSGEVRLNGRPLADwSPAELARRRAV 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALI------GGPP 2066
Cdd:PRK13548 81 LPQHSSLSFPFTVEEVVAMGRAPHGLSRAEDDALVAAALAQVDLAHLAGRDYPQLSGGEQQRVQLARVLAqlwepdGPPR 160
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2067 VVFLDEPTTGMDPK--------ARRFLWNCALSIVkegrsVVLtsHSMEECEALCTRMAIMVNGRFRCLGSVQ 2131
Cdd:PRK13548 161 WLLLDEPTSALDLAhqhhvlrlARQLAHERGLAVI-----VVL--HDLNLAARYADRIVLLHQGRLVADGTPA 226
|
|
| ABC_CysA_sulfate_importer |
cd03296 |
ATP-binding cassette domain of the sulfate transporter; Part of the ABC transporter complex ... |
1914-2129 |
3.80e-17 |
|
ATP-binding cassette domain of the sulfate transporter; Part of the ABC transporter complex cysAWTP involved in sulfate import. Responsible for energy coupling to the transport system. The complex is composed of two ATP-binding proteins (cysA), two transmembrane proteins (cysT and cysW), and a solute-binding protein (cysP). ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213263 [Multi-domain] Cd Length: 239 Bit Score: 83.16 E-value: 3.80e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIHEVHQNMGYC 1993
Cdd:cd03296 3 IEVRNVSKRF--GDFVALDDVSLDIPSGELVALLGPSGSGKTTLLRLIAGLERPDSGTILFGGEDA-TDVPVQERNVGFV 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGV----PEKEV-GKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:cd03296 80 FQHYALFRHMTVFDNVAFGLRVKPRserpPEAEIrAKVHE-LLKLVQLDWLADRYPAQLSGGQRQRVALARALAVEPKVL 158
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGS 2129
Cdd:cd03296 159 LLDEPFGALDAKVRKELRRWLRRLHDElHVTTVFVTHDQEEALEVADRVVVMNKGRIEQVGT 220
|
|
| 3a01204 |
TIGR00955 |
The Eye Pigment Precursor Transporter (EPP) Family protein; [Transport and binding proteins, ... |
923-1114 |
4.14e-17 |
|
The Eye Pigment Precursor Transporter (EPP) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273361 [Multi-domain] Cd Length: 617 Bit Score: 87.80 E-value: 4.14e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 923 FYEGQITSFLGHNGAGKTTTMSILTGLFPP---TSGTAYILGKDIRSEMssIRQNLGVCPQHNVLFDMLTVEEHIWFYAR 999
Cdd:TIGR00955 48 AKPGELLAVMGSSGAGKTTLMNALAFRSPKgvkGSGSVLLNGMPIDAKE--MRAISAYVQQDDLFIPTLTVREHLMFQAH 125
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1000 LK---GLSEKHVKAEMEQMALDVGL---------PPSKLKSktsqLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYS 1067
Cdd:TIGR00955 126 LRmprRVTKKEKRERVDEVLQALGLrkcantrigVPGRVKG----LSGGERKRLAFASELLTDPPLLFCDEPTSGLDSFM 201
|
170 180 190 200
....*....|....*....|....*....|....*....|....*....
gi 568925948 1068 RRGIWELLLKYRQGRTIILSTHHMDEADI--LGDRIAIISHGKLCCVGS 1114
Cdd:TIGR00955 202 AYSVVQVLKGLAQKGKTIICTIHQPSSELfeLFDKIILMAEGRVAYLGS 250
|
|
| ABC_ModC_like |
cd03299 |
ATP-binding cassette domain similar to the molybdate transporter; Archaeal protein closely ... |
1914-2131 |
4.17e-17 |
|
ATP-binding cassette domain similar to the molybdate transporter; Archaeal protein closely related to ModC. ModC is an ABC-type transporter and the ATPase component of a molybdate transport system that also includes the periplasmic binding protein ModA and the membrane protein ModB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213266 [Multi-domain] Cd Length: 235 Bit Score: 82.77 E-value: 4.17e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpaVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIHEVHQNMGYC 1993
Cdd:cd03299 1 LKVENLSKDWKEFK---LKNVSLEVERGDYFVILGPTGSGKSVLLETIAGFIKPDSGKILLNGKDI-TNLPPEKRDISYV 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVG-KVGEWAiRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDE 2072
Cdd:cd03299 77 PQNYALFPHMTVYKNIAYGLKKRKVDKKEIErKVLEIA-EMLGIDHLLNRKPETLSGGEQQRVAIARALVVNPKILLLDE 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2073 PTTGMDPKARRFLWNCALSIVKEGRSVVL-TSHSMEECEALCTRMAIMVNGRFRCLGSVQ 2131
Cdd:cd03299 156 PFSALDVRTKEKLREELKKIRKEFGVTVLhVTHDFEEAWALADKVAIMLNGKLIQVGKPE 215
|
|
| ABCC_Hemolysin |
cd03252 |
ATP-binding cassette domain of hemolysin B, subfamily C; The ABC-transporter hemolysin B is a ... |
908-1128 |
5.10e-17 |
|
ATP-binding cassette domain of hemolysin B, subfamily C; The ABC-transporter hemolysin B is a central component of the secretion machinery that translocates the toxin, hemolysin A, in a Sec-independent fashion across both membranes of E. coli. The hemolysin A (HlyA) transport machinery is composed of the ATP-binding cassette (ABC) transporter HlyB located in the inner membrane, hemolysin D (HlyD), also anchored in the inner membrane, and TolC, which resides in the outer membrane. HlyD apparently forms a continuous channel that bridges the entire periplasm, interacting with TolC and HlyB. This arrangement prevents the appearance of periplasmic intermediates of HlyA during substrate transport. Little is known about the molecular details of HlyA transport, but it is evident that ATP-hydrolysis by the ABC-transporter HlyB is a necessary source of energy.
Pssm-ID: 213219 [Multi-domain] Cd Length: 237 Bit Score: 82.53 E-value: 5.10e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFD 986
Cdd:cd03252 10 YKPDGPVILDNISLRIKPGEVVGIVGRSGSGKSTLTKLIQRFYVPENGRVLVDGHDLALaDPAWLRRQVGVVLQENVLFN 89
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 MLTVE-----------EHIWFYARLKGLSE--KHVKAEMEQMALDVGlppsklksktSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:cd03252 90 RSIRDnialadpgmsmERVIEAAKLAGAHDfiSELPEGYDTIVGEQG----------AGLSGGQRQRIAIARALIHNPRI 159
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMdEADILGDRIAIISHGKLCCVGSSLFLKNQLGTGYYL 1128
Cdd:cd03252 160 LIFDEATSALDYESEHAIMRNMHDICAGRTVIIIAHRL-STVKNADRIIVMEKGRIVEQGSHDELLAENGLYAYL 233
|
|
| PstB |
COG1117 |
ABC-type phosphate transport system, ATPase component [Inorganic ion transport and metabolism]; ... |
888-1109 |
5.57e-17 |
|
ABC-type phosphate transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 440734 [Multi-domain] Cd Length: 258 Bit Score: 83.16 E-value: 5.57e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 888 MEEEPTHLRLGVSIQNLvKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTT---TMSILTGLFPP--TSGTAYILGK 962
Cdd:COG1117 1 MTAPASTLEPKIEVRNL-NVYYGD-KQALKDINLDIPENKVTALIGPSGCGKSTllrCLNRMNDLIPGarVEGEILLDGE 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 963 DIRS---EMSSIRQNLGVCPQHNVLFDMlTVEEHIWFYARLKGLSEKHVKAEMEQMAL-DVGLPP---SKLKSKTSQLSG 1035
Cdd:COG1117 79 DIYDpdvDVVELRRRVGMVFQKPNPFPK-SIYDNVAYGLRLHGIKSKSELDEIVEESLrKAALWDevkDRLKKSALGLSG 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1036 GMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG1117 158 GQQQRLCIARALAVEPEVLLMDEPTSALDPISTAKIEELILELKKDYTIVIVTHNMQQAARVSDYTAFFYLGEL 231
|
|
| PRK14247 |
PRK14247 |
phosphate ABC transporter ATP-binding protein; Provisional |
916-1115 |
6.62e-17 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172735 [Multi-domain] Cd Length: 250 Bit Score: 82.65 E-value: 6.62e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLF-----PPTSGTAYILGKDI-RSEMSSIRQNLGVCPQHNVLFDMLT 989
Cdd:PRK14247 19 LDGVNLEIPDNTITALMGPSGSGKSTLLRVFNRLIelypeARVSGEVYLDGQDIfKMDVIELRRRVQMVFQIPNPIPNLS 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 990 VEEHIWFYARLKGLS------EKHVKAEMEQMAL--DVglpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTA 1061
Cdd:PRK14247 99 IFENVALGLKLNRLVkskkelQERVRWALEKAQLwdEV---KDRLDAPAGKLSGGQQQRLCIARALAFQPEVLLADEPTA 175
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1062 GVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS 1115
Cdd:PRK14247 176 NLDPENTAKIESLFLELKKDMTIVLVTHFPQQAARISDYVAFLYKGQIVEWGPT 229
|
|
| livF |
PRK11614 |
high-affinity branched-chain amino acid ABC transporter ATP-binding protein LivF; |
915-1109 |
6.70e-17 |
|
high-affinity branched-chain amino acid ABC transporter ATP-binding protein LivF;
Pssm-ID: 183231 [Multi-domain] Cd Length: 237 Bit Score: 82.23 E-value: 6.70e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSS--IRQNLGVCPQHNVLFDMLTVEE 992
Cdd:PRK11614 20 ALHEVSLHINQGEIVTLIGANGAGKTTLLGTLCGDPRATSGRIVFDGKDITDWQTAkiMREAVAIVPEGRRVFSRMTVEE 99
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 993 HI---WFYArlkglSEKHVKAEMEQMaldVGLPPsKLKSKTSQ----LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:PRK11614 100 NLamgGFFA-----ERDQFQERIKWV---YELFP-RLHERRIQragtMSGGEQQMLAIGRALMSQPRLLLLDEPSLGLAP 170
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 1066 YSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK11614 171 IIIQQIFDTIEQLREqGMTIFLVEQNANQALKLADRGYVLENGHV 215
|
|
| araG |
PRK11288 |
L-arabinose ABC transporter ATP-binding protein AraG; |
911-1109 |
7.85e-17 |
|
L-arabinose ABC transporter ATP-binding protein AraG;
Pssm-ID: 183077 [Multi-domain] Cd Length: 501 Bit Score: 86.12 E-value: 7.85e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR--SEMSSIRQNLGVCPQHNVLFDML 988
Cdd:PRK11288 16 GVK-ALDDISFDCRAGQVHALMGENGAGKSTLLKILSGNYQPDAGSILIDGQEMRfaSTTAALAAGVAIIYQELHLVPEM 94
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIW---FYARL----KGLSEKHVKAEMEQMALDVGlPPSKLKSktsqLSGGMQRKLSVALAFVGGSKVVILDEPTA 1061
Cdd:PRK11288 95 TVAENLYlgqLPHKGgivnRRLLNYEAREQLEHLGVDID-PDTPLKY----LSIGQRQMVEIAKALARNARVIAFDEPTS 169
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1062 GVdpySRRGIwELLLKY-----RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK11288 170 SL---SAREI-EQLFRVirelrAEGRVILYVSHRMEEIFALCDAITVFKDGRY 218
|
|
| PRK11160 |
PRK11160 |
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed |
878-1127 |
8.89e-17 |
|
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed
Pssm-ID: 236865 [Multi-domain] Cd Length: 574 Bit Score: 86.42 E-value: 8.89e-17
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 878 SSQKGVSEIcMEEEP------THL----RLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILT 947
Cdd:PRK11160 309 ASARRINEI-TEQKPevtfptTSTaaadQVSLTLNNVSFTYPDQPQPVLKGLSLQIKAGEKVALLGRTGCGKSTLLQLLT 387
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 948 GLFPPTSGTAYILGKDIR--SEmSSIRQNLGVCPQHNVLFDMlTVEEHiwfyarLKGLSEKHVKAEMEQMALDVGLppSK 1025
Cdd:PRK11160 388 RAWDPQQGEILLNGQPIAdySE-AALRQAISVVSQRVHLFSA-TLRDN------LLLAAPNASDEALIEVLQQVGL--EK 457
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1026 LKSKTS-----------QLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTH----- 1089
Cdd:PRK11160 458 LLEDDKglnawlgeggrQLSGGEQRRLGIARALLHDAPLLLLDEPTEGLDAETERQILELLAEHAQNKTVLMITHrltgl 537
|
250 260 270
....*....|....*....|....*....|....*....
gi 568925948 1090 -HMdeadilgDRIAIISHGKLCCVGSSLFLKNQLGtGYY 1127
Cdd:PRK11160 538 eQF-------DRICVMDNGQIIEQGTHQELLAQQG-RYY 568
|
|
| ABCC_Protease_Secretion |
cd03246 |
ATP-binding cassette domain of PrtD, subfamily C; This family represents the ABC component of ... |
900-1109 |
1.19e-16 |
|
ATP-binding cassette domain of PrtD, subfamily C; This family represents the ABC component of the protease secretion system PrtD, a 60-kDa integral membrane protein sharing 37% identity with HlyB, the ABC component of the alpha-hemolysin secretion pathway, in the C-terminal domain. They export degradative enzymes by using a type I protein secretion system and lack an N-terminal signal peptide, but contain a C-terminal secretion signal. The Type I secretion apparatus is made up of three components, an ABC transporter, a membrane fusion protein (MFP), and an outer membrane protein (OMP). For the HlyA transporter complex, HlyB (ABC transporter) and HlyD (MFP) reside in the inner membrane of E. coli. The OMP component is TolC, which is thought to interact with the MFP to form a continuous channel across the periplasm from the cytoplasm to the exterior. HlyB belongs to the family of ABC transporters, which are ubiquitous, ATP-dependent transmembrane pumps or channels. The spectrum of transport substrates ranges from inorganic ions, nutrients such as amino acids, sugars, or peptides, hydrophobic drugs, to large polypeptides, such as HlyA.
Pssm-ID: 213213 [Multi-domain] Cd Length: 173 Bit Score: 79.57 E-value: 1.19e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVC 978
Cdd:cd03246 2 EVENVSFRYPGAEPPVLRNVSFSIEPGESLAIIGPSGSGKSTLARLILGLLRPTSGRVRLDGADISQwDPNELGDHVGYL 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMlTVEEHIwfyarlkglsekhvkaemeqmaldvglppsklksktsqLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03246 82 PQDDELFSG-SIAENI--------------------------------------LSGGQRQRLGLARALYGNPRILVLDE 122
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMdEADILGDRIAIISHGKL 1109
Cdd:cd03246 123 PNSHLDVEGERALNQAIAALKaAGATRIVIAHRP-ETLASADRILVLEDGRV 173
|
|
| PRK14267 |
PRK14267 |
phosphate ABC transporter ATP-binding protein; Provisional |
912-1113 |
1.23e-16 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 184596 [Multi-domain] Cd Length: 253 Bit Score: 81.81 E-value: 1.23e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 912 MKVAVDGLALNFYEGQ---------------ITSFLGHNGAGKTTTMSILTGLF-----PPTSGTAYILGKDIRSEMSS- 970
Cdd:PRK14267 1 MKFAIETVNLRVYYGSnhvikgvdlkipqngVFALMGPSGCGKSTLLRTFNRLLelneeARVEGEVRLFGRNIYSPDVDp 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 --IRQNLGVCPQHNVLFDMLTVEEHIWFYARLKGLS------EKHVKAEMEQMAL--DVglpPSKLKSKTSQLSGGMQRK 1040
Cdd:PRK14267 81 ieVRREVGMVFQYPNPFPHLTIYDNVAIGVKLNGLVkskkelDERVEWALKKAALwdEV---KDRLNDYPSNLSGGQRQR 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1041 LSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:PRK14267 158 LVIARALAMKPKILLMDEPTANIDPVGTAKIEELLFELKKEYTIVLVTHSPAQAARVSDYVAFLYLGKLIEVG 230
|
|
| ABC_PstB_phosphate_transporter |
cd03260 |
ATP-binding cassette domain of the phosphate transport system; Phosphate uptake is of ... |
1914-2123 |
1.37e-16 |
|
ATP-binding cassette domain of the phosphate transport system; Phosphate uptake is of fundamental importance in the cell physiology of bacteria because phosphate is required as a nutrient. The Pst system of E. coli comprises four distinct subunits encoded by the pstS, pstA, pstB, and pstC genes. The PstS protein is a phosphate-binding protein located in the periplasmic space. PstA and PstC are hydrophobic and they form the transmembrane portion of the Pst system. PstB is the catalytic subunit, which couples the energy of ATP hydrolysis to the import of phosphate across cellular membranes through the Pst system, often referred as ABC-protein. PstB belongs to one of the largest superfamilies of proteins characterized by a highly conserved adenosine triphosphate (ATP) binding cassette (ABC), which is also a nucleotide binding domain (NBD).
Pssm-ID: 213227 [Multi-domain] Cd Length: 227 Bit Score: 81.07 E-value: 1.37e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG------DTPVTrGDAFL---NKNSILSNIH 1984
Cdd:cd03260 1 IELRDLNVYYGDKH--ALKDISLDIPKGEITALIGPSGCGKSTLLRLLNRlndlipGAPDE-GEVLLdgkDIYDLDVDVL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVHQNMGYCPQ----FDAitellTGREHVEFFALLRGV-PEKEVGKVGEWAIRKLGLVKYGEK--YASNYSGGNKRKLST 2057
Cdd:cd03260 78 ELRRRVGMVFQkpnpFPG-----SIYDNVAYGLRLHGIkLKEELDERVEEALRKAALWDEVKDrlHALGLSGGQQQRLCL 152
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEgRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03260 153 ARALANEPEVLLLDEPTSALDPISTAKIEELIAELKKE-YTIVIVTHNMQQAARVADRTAFLLNGR 217
|
|
| xylG |
TIGR02633 |
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose ... |
916-1110 |
1.50e-16 |
|
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose isomerase and xylulokinase enzymes for xylose utilization. Members of this protein family are the ATP-binding cassette (ABC) subunit of the known or predicted high-affinity xylose ABC transporter for xylose import. These genes, which closely resemble other sugar transport ABC transporter genes, typically are encoded near xylose utilization enzymes and regulatory proteins. Note that this form of the transporter contains two copies of the ABC transporter domain (pfam00005). [Transport and binding proteins, Carbohydrates, organic alcohols, and acids]
Pssm-ID: 131681 [Multi-domain] Cd Length: 500 Bit Score: 85.26 E-value: 1.50e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPT-SGTAYILGK--DIRSEMSSIRQNLGVCPQ----HNVLFDMl 988
Cdd:TIGR02633 276 VDDVSFSLRRGEILGVAGLVGAGRTELVQALFGAYPGKfEGNVFINGKpvDIRNPAQAIRAGIAMVPEdrkrHGIVPIL- 354
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWFyARLKGLSEK-HVKAEMEQMALDVGLppSKLKSKTS-------QLSGGMQRKLSVALAFVGGSKVVILDEPT 1060
Cdd:TIGR02633 355 GVGKNITL-SVLKSFCFKmRIDAAAELQIIGSAI--QRLKVKTAspflpigRLSGGNQQKAVLAKMLLTNPRVLILDEPT 431
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1061 AGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKLC 1110
Cdd:TIGR02633 432 RGVDVGAKYEIYKLINQLaQEGVAIIVVSSELAEVLGLSDRVLVIGEGKLK 482
|
|
| PRK10070 |
PRK10070 |
proline/glycine betaine ABC transporter ATP-binding protein ProV; |
1936-2141 |
1.80e-16 |
|
proline/glycine betaine ABC transporter ATP-binding protein ProV;
Pssm-ID: 182221 [Multi-domain] Cd Length: 400 Bit Score: 84.31 E-value: 1.80e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1936 IGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL----SNIHEVH-QNMGYCPQFDAITELLTGREHVE 2010
Cdd:PRK10070 49 LAIEEGEIFVIMGLSGSGKSTMVRLLNRLIEPTRGQVLIDGVDIAkisdAELREVRrKKIAMVFQSFALMPHMTVLDNTA 128
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2011 FFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCAL 2090
Cdd:PRK10070 129 FGMELAGINAEERREKALDALRQVGLENYAHSYPDELSGGMRQRVGLARALAINPDILLMDEAFSALDPLIRTEMQDELV 208
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2091 SI-VKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGDGY 2141
Cdd:PRK10070 209 KLqAKHQRTIVFISHDLDEAMRIGDRIAIMQNGEVVQVGTPDEILNNPANDY 260
|
|
| ABC_FtsE |
cd03292 |
Cell division ATP-binding protein FtsE; The FtsEX complex resembles an ABC transporter, where ... |
1914-2123 |
1.81e-16 |
|
Cell division ATP-binding protein FtsE; The FtsEX complex resembles an ABC transporter, where FtsE is the ATPase and the membrane subunit FtsX resembles a permease subunit. But rather than transporting any substrate, the complex acts in cell division by undergoing conformational changes that alter the activity of cell wall hydrolases located outside the plasma membrane. The complex is widely conserved in bacteria, but also extremely divergent in sequence between different lineages
Pssm-ID: 213259 [Multi-domain] Cd Length: 214 Bit Score: 80.53 E-value: 1.81e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKrKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSiLSNIHE-----VHQ 1988
Cdd:cd03292 1 IEFINVTKTYPNG-TAALDGINISISAGEFVFLVGPSGAGKSTLLKLIYKEELPTSGTIRVNGQD-VSDLRGraipyLRR 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:cd03292 79 KIGVVFQDFRLLPDRNVYENVAFALEVTGVPPREIRKRVPAALELVGLSHKHRALPAELSGGEQQRVAIARAIVNSPTIL 158
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03292 159 IADEPTGNLDPDTTWEIMNLLKKINKAGTTVVVATHAKELVDTTRHRVIALERGK 213
|
|
| CydC |
TIGR02868 |
thiol reductant ABC exporter, CydC subunit; The gene pair cydCD encodes an ABC-family ... |
1929-2105 |
3.20e-16 |
|
thiol reductant ABC exporter, CydC subunit; The gene pair cydCD encodes an ABC-family transporter in which each gene contains an N-terminal membrane-spanning domain (pfam00664) and a C-terminal ATP-binding domain (pfam00005). In E. coli these genes were discovered as mutants which caused the terminal heme-copper oxidase complex cytochrome bd to fail to assemble. Recent work has shown that the transporter is involved in export of redox-active thiol compounds such as cysteine and glutathione. The linkage to assembly of the cytochrome bd complex is further supported by the conserved operon structure found outside the gammaproteobacteria (cydABCD) containing both the transporter and oxidase genes components. The genes used as the seed members for this model are all either found in the gammproteobacterial context or the CydABCD context. All members of this family scoring above trusted at the time of its creation were from genomes which encode a cytochrome bd complex.
Pssm-ID: 274331 [Multi-domain] Cd Length: 530 Bit Score: 84.33 E-value: 3.20e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIH-EVHQNMGYCPQ----FDA-ITE- 2001
Cdd:TIGR02868 349 PVLDGVSLDLPPGERVAILGPSGSGKSTLLATLAGLLDPLQGEVTLDGVPVSSLDQdEVRRRVSVCAQdahlFDTtVREn 428
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2002 LLTGREHV---EFFALLRGVpekevgKVGEWaIRKL--GL-VKYGEKYASnYSGGNKRKLSTAMALIGGPPVVFLDEPTT 2075
Cdd:TIGR02868 429 LRLARPDAtdeELWAALERV------GLADW-LRALpdGLdTVLGEGGAR-LSGGERQRLALARALLADAPILLLDEPTE 500
|
170 180 190
....*....|....*....|....*....|
gi 568925948 2076 GMDPKARRFLWNcALSIVKEGRSVVLTSHS 2105
Cdd:TIGR02868 501 HLDAETADELLE-DLLAALSGRTVVLITHH 529
|
|
| CP_lyasePhnK |
TIGR02323 |
phosphonate C-P lyase system protein PhnK; Members of this family are the PhnK protein of C-P ... |
899-1109 |
4.63e-16 |
|
phosphonate C-P lyase system protein PhnK; Members of this family are the PhnK protein of C-P lyase systems for utilization of phosphonates. These systems resemble phosphonatase-based systems in having a three component ABC transporter, where TIGR01097 is the permease, TIGR01098 is the phosphonates binding protein, and TIGR02315 is the ATP-binding cassette (ABC) protein. They differ, however, in having, typically, ten or more additional genes, many of which are believed to form a membrane-associated complex. This protein (PhnK) and the adjacent-encoded PhnL resemble transporter ATP-binding proteins but are suggested, based on mutatgenesis studies, to be part of this complex rather than part of a transporter per se. [Central intermediary metabolism, Phosphorus compounds]
Pssm-ID: 188208 [Multi-domain] Cd Length: 253 Bit Score: 80.26 E-value: 4.63e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS----EMSS---- 970
Cdd:TIGR02323 4 LQVSGLSKSY--GGGKGCRDVSFDLYPGEVLGIVGESGSGKSTLLGCLAGRLAPDHGTATYIMRSGAElelyQLSEaerr 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 --IRQNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKH---VKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVAL 1045
Cdd:TIGR02323 82 rlMRTEWGFVHQNPRDGLRMRVSAGANIGERLMAIGARHygnIRATAQDWLEEVEIDPTRIDDLPRAFSGGMQQRLQIAR 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1046 AFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:TIGR02323 162 NLVTRPRLVFMDEPTGGLDVSVQARLLDLLrgLVRDLGLAVIIVTHDLGVARLLAQRLLVMQQGRV 227
|
|
| cbiO |
PRK13638 |
energy-coupling factor ABC transporter ATP-binding protein; |
918-1114 |
4.72e-16 |
|
energy-coupling factor ABC transporter ATP-binding protein;
Pssm-ID: 184198 [Multi-domain] Cd Length: 271 Bit Score: 80.82 E-value: 4.72e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 918 GLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI---RSEMSSIRQNLGVC---PQHNVLFDmlTVE 991
Cdd:PRK13638 19 GLNLDFSLSPVTGLVGANGCGKSTLFMNLSGLLRPQKGAVLWQGKPLdysKRGLLALRQQVATVfqdPEQQIFYT--DID 96
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 992 EHIWFYARLKGLSEKHVKAEMEQmALDVgLPPSKLKSKTSQ-LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRG 1070
Cdd:PRK13638 97 SDIAFSLRNLGVPEAEITRRVDE-ALTL-VDAQHFRHQPIQcLSHGQKKRVAIAGALVLQARYLLLDEPTAGLDPAGRTQ 174
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 1071 IWELLLK-YRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK13638 175 MIAIIRRiVAQGNHVIISSHDIDLIYEISDAVYVLRQGQILTHGA 219
|
|
| lolD |
PRK11629 |
lipoprotein-releasing ABC transporter ATP-binding protein LolD; |
903-1098 |
5.87e-16 |
|
lipoprotein-releasing ABC transporter ATP-binding protein LolD;
Pssm-ID: 183244 [Multi-domain] Cd Length: 233 Bit Score: 79.47 E-value: 5.87e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 903 NLVKVYRDGmKVAVDGL---ALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSS-----IR-Q 973
Cdd:PRK11629 10 NLCKRYQEG-SVQTDVLhnvSFSIGEGEMMAIVGSSGSGKSTLLHLLGGLDTPTSGDVIFNGQPM-SKLSSaakaeLRnQ 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 NLGVCPQ-HNVLFDmLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:PRK11629 88 KLGFIYQfHHLLPD-FTALENVAMPLLIGKKKPAEINSRALEMLAAVGLE-HRANHRPSELSGGERQRVAIARALVNNPR 165
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILG 1098
Cdd:PRK11629 166 LVLADEPTGNLDARNADSIFQLLgeLNRLQGTAFLVVTHDLQLAKRMS 213
|
|
| cbiO |
PRK13647 |
cobalt transporter ATP-binding subunit; Provisional |
1911-2136 |
6.35e-16 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237457 [Multi-domain] Cd Length: 274 Bit Score: 80.16 E-value: 6.35e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHEVHQN 1989
Cdd:PRK13647 2 DNIIEVEDLHFRYKDGTK-ALKGLSLSIPEGSKTALLGPNGAGKSTLLLHLNGIYLPQRGRVKVMGREVnAENEKWVRSK 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQfDAITELLTGR--EHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:PRK13647 81 VGLVFQ-DPDDQVFSSTvwDDVAFGPVNMGLDKDEVERRVEEALKAVRMWDFRDKPPYHLSYGQKKRVAIAGVLAMDPDV 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNR 2136
Cdd:PRK13647 160 IVLDEPMAYLDPRGQETLMEILDRLHNQGKTVIVATHDVDLAAEWADQVIVLKEGRVLAEGDKSLLTDE 228
|
|
| PRK10762 |
PRK10762 |
D-ribose transporter ATP binding protein; Provisional |
916-1110 |
7.33e-16 |
|
D-ribose transporter ATP binding protein; Provisional
Pssm-ID: 236755 [Multi-domain] Cd Length: 501 Bit Score: 83.13 E-value: 7.33e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSemssirqnlgVCPQ--------------- 980
Cdd:PRK10762 268 VNDVSFTLRKGEILGVSGLMGAGRTELMKVLYGALPRTSGYVTLDGHEVVT----------RSPQdglangivyisedrk 337
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLFDMLTVEEHIWFYArLKGLSEK--HVKAEMEQMALD--VGLPPSKLKSKTSQ---LSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK10762 338 RDGLVLGMSVKENMSLTA-LRYFSRAggSLKHADEQQAVSdfIRLFNIKTPSMEQAiglLSGGNQQKVAIARGLMTRPKV 416
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEadILG--DRIAIISHGKLC 1110
Cdd:PRK10762 417 LILDEPTRGVDVGAKKEIYQLINQFKAeGLSIILVSSEMPE--VLGmsDRILVMHEGRIS 474
|
|
| cbiO |
PRK13632 |
cobalt transporter ATP-binding subunit; Provisional |
1915-2123 |
7.46e-16 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237452 [Multi-domain] Cd Length: 271 Bit Score: 80.03 E-value: 7.46e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFL--------NKNSILSNIHEV 1986
Cdd:PRK13632 9 KVENVSFSYPNSENNALKNVSFEINEGEYVAILGHNGSGKSTISKILTGLLKPQSGEIKIdgitiskeNLKEIRKKIGII 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNmgycP--QFDAITelltgrehVE---FFALL-RGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:PRK13632 89 FQN----PdnQFIGAT--------VEddiAFGLEnKKVPPKKMKDIIDDLAKKVGMEDYLDKEPQNLSGGQKQRVAIASV 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLT-SHSMEECeALCTRMAIMVNGR 2123
Cdd:PRK13632 157 LALNPEIIIFDESTSMLDPKGKREIKKIMVDLRKTRKKTLISiTHDMDEA-ILADKVIVFSEGK 219
|
|
| PRK10762 |
PRK10762 |
D-ribose transporter ATP binding protein; Provisional |
911-1108 |
7.72e-16 |
|
D-ribose transporter ATP binding protein; Provisional
Pssm-ID: 236755 [Multi-domain] Cd Length: 501 Bit Score: 83.13 E-value: 7.72e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR---------SEMSSIRQNLGVCPQh 981
Cdd:PRK10762 16 GVK-ALSGAALNVYPGRVMALVGENGAGKSTMMKVLTGIYTRDAGSILYLGKEVTfngpkssqeAGIGIIHQELNLIPQ- 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 982 nvlfdmLTVEEHIW----FYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILD 1057
Cdd:PRK10762 94 ------LTIAENIFlgreFVNRFGRIDWKKMYAEADKLLARLNLRFSS-DKLVGELSIGEQQMVEIAKVLSFESKVIIMD 166
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1058 EPT-AGVDPYSR---RGIWELllkYRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK10762 167 EPTdALTDTETEslfRVIREL---KSQGRGIVYISHRLKEIFEICDDVTVFRDGQ 218
|
|
| PRK14243 |
PRK14243 |
phosphate transporter ATP-binding protein; Provisional |
902-1102 |
7.96e-16 |
|
phosphate transporter ATP-binding protein; Provisional
Pssm-ID: 184588 [Multi-domain] Cd Length: 264 Bit Score: 79.83 E-value: 7.96e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 902 QNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSI---LTGLFPP--TSGTAYILGKDI---RSEMSSIRQ 973
Cdd:PRK14243 14 ENLNVYY--GSFLAVKNVWLDIPKNQITAFIGPSGCGKSTILRCfnrLNDLIPGfrVEGKVTFHGKNLyapDVDPVEVRR 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 NLGVCPQHNVLFDMlTVEEHIWFYARL---KGLSEKHVKAEMEQMAL--DVglpPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:PRK14243 92 RIGMVFQKPNPFPK-SIYDNIAYGARIngyKGDMDELVERSLRQAALwdEV---KDKLKQSGLSLSGGQQQRLCIARAIA 167
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIA 1102
Cdd:PRK14243 168 VQPEVILMDEPCSALDPISTLRIEELMHELKEQYTIIIVTHNMQQAARVSDMTA 221
|
|
| ABCG_PDR_domain2 |
cd03232 |
Second domain of the pleiotropic drug resistance-like (PDR) subfamily G of ATP-binding ... |
1938-2104 |
8.06e-16 |
|
Second domain of the pleiotropic drug resistance-like (PDR) subfamily G of ATP-binding cassette transporters; The pleiotropic drug resistance (PDR) is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. This PDR subfamily represents domain I of its (ABC-IM)2 organization. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds including sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213199 [Multi-domain] Cd Length: 192 Bit Score: 78.05 E-value: 8.06e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTG--DTPVTRGDAFLNKNSILSNIHEVhqnMGYCPQFDAITELLTGREHVEFFALL 2015
Cdd:cd03232 30 VKPGTLTALMGESGAGKTTLLDVLAGrkTAGVITGEILINGRPLDKNFQRS---TGYVEQQDVHSPNLTVREALRFSALL 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2016 RGVpekevgkvgewairklglvkygekyasnySGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKAR----RFLWNCALS 2091
Cdd:cd03232 107 RGL-----------------------------SVEQRKRLTIGVELAAKPSILFLDEPTSGLDSQAAynivRFLKKLADS 157
|
170
....*....|...
gi 568925948 2092 ivkeGRSVVLTSH 2104
Cdd:cd03232 158 ----GQAILCTIH 166
|
|
| PRK09700 |
PRK09700 |
D-allose ABC transporter ATP-binding protein AlsA; |
1930-2135 |
8.31e-16 |
|
D-allose ABC transporter ATP-binding protein AlsA;
Pssm-ID: 182036 [Multi-domain] Cd Length: 510 Bit Score: 82.91 E-value: 8.31e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN---------KNSILSNIHEVHQNMGYCPQFDAIT 2000
Cdd:PRK09700 20 ALKSVNLTVYPGEIHALLGENGAGKSTLMKVLSGIHEPTKGTITINninynkldhKLAAQLGIGIIYQELSVIDELTVLE 99
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 ELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPK 2080
Cdd:PRK09700 100 NLYIGRHLTKKVCGVNIIDWREMRVRAAMMLLRVGLKVDLDEKVANLSISHKQMLEIAKTLMLDAKVIIMDEPTSSLTNK 179
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2081 ARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:PRK09700 180 EVDYLFLIMNQLRKEGTAIVYISHKLAEIRRICDRYTVMKDGSSVCSGMVSDVSN 234
|
|
| CydD |
COG4988 |
ABC-type transport system involved in cytochrome bd biosynthesis, ATPase and permease ... |
1892-2133 |
8.63e-16 |
|
ABC-type transport system involved in cytochrome bd biosynthesis, ATPase and permease components [Energy production and conversion, Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 444012 [Multi-domain] Cd Length: 563 Bit Score: 83.27 E-value: 8.63e-16
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1892 DEDEDVRRERQRILDGGGQNDIlEIKELTKIYRrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGD 1971
Cdd:COG4988 316 DAPEPAAPAGTAPLPAAGPPSI-ELEDVSFSYP-GGRPALDGLSLTIPPGERVALVGPSGAGKSTLLNLLLGFLPPYSGS 393
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1972 AFLNKNSILS-NIHEVHQNMGYCPQFdaiTELLTG--REHVEFFAllRGVPEKEVgkvgEWAIRKLGL---VKY------ 2039
Cdd:COG4988 394 ILINGVDLSDlDPASWRRQIAWVPQN---PYLFAGtiRENLRLGR--PDASDEEL----EAALEAAGLdefVAAlpdgld 464
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2040 ---GEKyASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSHSMEECeALCTRM 2116
Cdd:COG4988 465 tplGEG-GRGLSGGQAQRLALARALLRDAPLLLLDEPTAHLDAETEAEILQ-ALRRLAKGRTVILITHRLALL-AQADRI 541
|
250
....*....|....*..
gi 568925948 2117 AIMVNGRFRCLGSVQHL 2133
Cdd:COG4988 542 LVLDDGRIVEQGTHEEL 558
|
|
| glnQ |
PRK09493 |
glutamine ABC transporter ATP-binding protein GlnQ; |
1913-2123 |
1.05e-15 |
|
glutamine ABC transporter ATP-binding protein GlnQ;
Pssm-ID: 181906 [Multi-domain] Cd Length: 240 Bit Score: 78.98 E-value: 1.05e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKiyRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS---NIHEVHQN 1989
Cdd:PRK09493 1 MIEFKNVSK--HFGPTQVLHNIDLNIDQGEVVVIIGPSGSGKSTLLRCINKLEEITSGDLIVDGLKVNDpkvDERLIRQE 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYC-PQFDAITELlTGREHVEFFAL-LRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPV 2067
Cdd:PRK09493 79 AGMVfQQFYLFPHL-TALENVMFGPLrVRGASKEEAEKQARELLAKVGLAERAHHYPSELSGGQQQRVAIARALAVKPKL 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2068 VFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK09493 158 MLFDEPTSALDPELRHEVLKVMQDLAEEGMTMVIVTHEIGFAEKVASRLIFIDKGR 213
|
|
| cbiO |
PRK13640 |
energy-coupling factor transporter ATPase; |
1911-2110 |
1.27e-15 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184200 [Multi-domain] Cd Length: 282 Bit Score: 79.46 E-value: 1.27e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG--------DTPVTRGDAFLNKNSILsN 1982
Cdd:PRK13640 3 DNIVEFKHVSFTYPDSKKPALNDISFSIPRGSWTALIGHNGSGKSTISKLINGlllpddnpNSKITVDGITLTAKTVW-D 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1983 IHE----VHQNmgycP--QFDAITElltgREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLS 2056
Cdd:PRK13640 82 IREkvgiVFQN----PdnQFVGATV----GDDVAFGLENRAVPRPEMIKIVRDVLADVGMLDYIDSEPANLSGGQKQRVA 153
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2057 TAMALIGGPPVVFLDEPTTGMDPKARrflwNCALSIVKE-----GRSVVLTSHSMEECE 2110
Cdd:PRK13640 154 IAGILAVEPKIIILDESTSMLDPAGK----EQILKLIRKlkkknNLTVISITHDIDEAN 208
|
|
| ABC_CcmA_heme_exporter |
cd03231 |
Cytochrome c biogenesis ATP-binding export protein; CcmA, the ATP-binding component of the ... |
1914-2104 |
1.32e-15 |
|
Cytochrome c biogenesis ATP-binding export protein; CcmA, the ATP-binding component of the bacterial CcmAB transporter. The CCM family is involved in bacterial cytochrome c biogenesis. Cytochrome c maturation in E. coli requires the ccm operon, which encodes eight membrane proteins (CcmABCDEFGH). CcmE is a periplasmic heme chaperon that binds heme covalently and transfers it onto apocytochrome c in the presence of CcmF, CcmG, and CcmH. The CcmAB proteins represent an ABC transporter and the CcmCD proteins participate in heme transfer to CcmE.
Pssm-ID: 213198 [Multi-domain] Cd Length: 201 Bit Score: 77.53 E-value: 1.32e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03231 1 LEADELT--CERDGRALFSGLSFTLAAGEALQVTGPNGSGKTTLLRILAGLSPPLAGRVLLNGGPLDFQRDSIARGLLYL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVgkvgewAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03231 79 GHAPGIKTTLSVLENLRFWHADHSDEQVEE------ALARVGLNGFEDRPVAQLSAGQQRRVALARLLLSGRPLWILDEP 152
|
170 180 190
....*....|....*....|....*....|....*.
gi 568925948 2074 TTGMDPK-----ARRFLWNCAlsivkEGRSVVLTSH 2104
Cdd:cd03231 153 TTALDKAgvarfAEAMAGHCA-----RGGMVVLTTH 183
|
|
| PRK10851 |
PRK10851 |
sulfate/thiosulfate ABC transporter ATP-binding protein CysA; |
899-1114 |
1.50e-15 |
|
sulfate/thiosulfate ABC transporter ATP-binding protein CysA;
Pssm-ID: 182778 [Multi-domain] Cd Length: 353 Bit Score: 80.51 E-value: 1.50e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrDGMKVAVDgLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGVC 978
Cdd:PRK10851 3 IEIANIKKSF-GRTQVLND-ISLDIPSGQMVALLGPSGSGKTTLLRIIAGLEHQTSGHIRFHGTDV-SRLHARDRKVGFV 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQHNVLFDMLTVEEHIWF----YARLKGLSEKHVKAEMEQMALDVGLppSKLKSK-TSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK10851 80 FQHYALFRHMTVFDNIAFgltvLPRRERPNAAAIKAKVTQLLEMVQL--AHLADRyPAQLSGGQKQRVALARALAVEPQI 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELL------LKYrqgrTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK10851 158 LLLDEPFGALDAQVRKELRRWLrqlheeLKF----TSVFVTHDQEEAMEVADRVVVMSQGNIEQAGT 220
|
|
| PRK13633 |
PRK13633 |
energy-coupling factor transporter ATPase; |
1911-2123 |
1.71e-15 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237453 [Multi-domain] Cd Length: 280 Bit Score: 79.36 E-value: 1.71e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYRR----KRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNK--NSILSNIH 1984
Cdd:PRK13633 2 NEMIKCKNVSYKYESneesTEKLALDDVNLEVKKGEFLVILGRNGSGKSTIAKHMNALLIPSEGKVYVDGldTSDEENLW 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVHQNMGYCPQ------FDAITElltgrEHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYgEKYASNY-SGGNKRKLST 2057
Cdd:PRK13633 82 DIRNKAGMVFQnpdnqiVATIVE-----EDVAFGPENLGIPPEEIRERVDESLKKVGMYEY-RRHAPHLlSGGQKQRVAI 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECeALCTRMAIMVNGR 2123
Cdd:PRK13633 156 AGILAMRPECIIFDEPTAMLDPSGRREVVNTIKELNKKyGITIILITHYMEEA-VEADRIIVMDSGK 221
|
|
| cbiO |
PRK13644 |
energy-coupling factor transporter ATPase; |
899-1114 |
2.25e-15 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 106587 [Multi-domain] Cd Length: 274 Bit Score: 78.88 E-value: 2.25e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI--RSEMSSIRQNLG 976
Cdd:PRK13644 2 IRLENVSYSYPDGTP-ALENINLVIKKGEYIGIIGKNGSGKSTLALHLNGLLRPQKGKVLVSGIDTgdFSKLQGIRKLVG 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQH-NVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSqLSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:PRK13644 81 IVFQNpETQFVGRTVEEDLAFGPENLCLPPIEIRKRVDRALAEIGLEKYRHRSPKT-LSGGQGQCVALAGILTMEPECLI 159
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1056 LDEPTAGVDPYSRRGIWELLLK-YRQGRTIILSTHHMDEADIlGDRIAIISHGKLCCVGS 1114
Cdd:PRK13644 160 FDEVTSMLDPDSGIAVLERIKKlHEKGKTIVYITHNLEELHD-ADRIIVMDRGKIVLEGE 218
|
|
| DppD |
COG0444 |
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid ... |
1913-2123 |
2.36e-15 |
|
ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component [Amino acid transport and metabolism, Inorganic ion transport and metabolism];
Pssm-ID: 440213 [Multi-domain] Cd Length: 320 Bit Score: 79.33 E-value: 2.36e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP---VTRGDAFLNKNSILS----NI 1983
Cdd:COG0444 1 LLEVRNLKVYFPTRRGVvkAVDGVSFDVRRGETLGLVGESGSGKSTLARAILGLLPppgITSGEILFDGEDLLKlsekEL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1984 HEV-HQNMGYCPQfDAITEL---LTGREHV-EFFALLRGVPEKEVGKVgewAIRKLGLVK--YGEKYASNY----SGGNK 2052
Cdd:COG0444 81 RKIrGREIQMIFQ-DPMTSLnpvMTVGDQIaEPLRIHGGLSKAEARER---AIELLERVGlpDPERRLDRYphelSGGMR 156
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2053 RKLSTAMALIGGPPVVFLDEPTTGMDpkarrflwncaLSI-----------VKE-GRSVVLTSHSMEECEALCTRMAIMV 2120
Cdd:COG0444 157 QRVMIARALALEPKLLIADEPTTALD-----------VTIqaqilnllkdlQRElGLAILFITHDLGVVAEIADRVAVMY 225
|
...
gi 568925948 2121 NGR 2123
Cdd:COG0444 226 AGR 228
|
|
| MglA |
COG1129 |
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism]; |
1913-2124 |
2.50e-15 |
|
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440745 [Multi-domain] Cd Length: 497 Bit Score: 81.22 E-value: 2.50e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrkrkPAV---DRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHE--- 1985
Cdd:COG1129 4 LLEMRGISKSF-----GGVkalDGVSLELRPGEVHALLGENGAGKSTLMKILSGVYQPDSGEILLDGEPVrFRSPRDaqa 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 -----VHQNMGYCPQFDaITE-LLTGREhveffallrgvpekevgkvgewaIRKLGLVKYGEKYA--------------- 2044
Cdd:COG1129 79 agiaiIHQELNLVPNLS-VAEnIFLGRE-----------------------PRRGGLIDWRAMRRrarellarlgldidp 134
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2045 ----SNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLwncaLSIVK----EGRSVVLTSHSMEECEALCTRM 2116
Cdd:COG1129 135 dtpvGDLSVAQQQLVEIARALSRDARVLILDEPTASLTEREVERL----FRIIRrlkaQGVAIIYISHRLDEVFEIADRV 210
|
....*...
gi 568925948 2117 AIMVNGRF 2124
Cdd:COG1129 211 TVLRDGRL 218
|
|
| metN |
PRK11153 |
DL-methionine transporter ATP-binding subunit; Provisional |
1915-2130 |
2.60e-15 |
|
DL-methionine transporter ATP-binding subunit; Provisional
Pssm-ID: 236863 [Multi-domain] Cd Length: 343 Bit Score: 79.84 E-value: 2.60e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI--LSN--IHEVHQ 1988
Cdd:PRK11153 3 ELKNISKVFPQGGRTihALNNVSLHIPAGEIFGVIGASGAGKSTLIRCINLLERPTSGRVLVDGQDLtaLSEkeLRKARR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQ-FDaiteLLTGREHVEFFAL---LRGVPEKEVGK-VGEWairkLGLVKYGEK---YASNYSGGNKRKLSTAMA 2060
Cdd:PRK11153 83 QIGMIFQhFN----LLSSRTVFDNVALpleLAGTPKAEIKArVTEL----LELVGLSDKadrYPAQLSGGQKQRVAIARA 154
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLwncaLSIVKE-----GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSV 2130
Cdd:PRK11153 155 LASNPKVLLCDEATSALDPATTRSI----LELLKDinrelGLTIVLITHEMDVVKRICDRVAVIDAGRLVEQGTV 225
|
|
| Uup |
COG0488 |
ATPase components of ABC transporters with duplicated ATPase domains [General function ... |
1928-2074 |
2.64e-15 |
|
ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only];
Pssm-ID: 440254 [Multi-domain] Cd Length: 520 Bit Score: 81.26 E-value: 2.64e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilSNIHEVHQNMGYCPQFDAITELLTGRE 2007
Cdd:COG0488 11 RPLLDDVSLSINPGDRIGLVGRNGAGKSTLLKILAGELEPDSGEVSIPKG---LRIGYLPQEPPLDDDLTVLDTVLDGDA 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 HV-----EFFALLRGVPE-----KEVGKV-------GEWAIR--------KLGL-VKYGEKYASNYSGGNKRKLSTAMAL 2061
Cdd:COG0488 88 ELraleaELEELEAKLAEpdedlERLAELqeefealGGWEAEaraeeilsGLGFpEEDLDRPVSELSGGWRRRVALARAL 167
|
170
....*....|...
gi 568925948 2062 IGGPPVVFLDEPT 2074
Cdd:COG0488 168 LSEPDLLLLDEPT 180
|
|
| PhnL |
COG4778 |
Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [Inorganic ion ... |
1913-2123 |
2.73e-15 |
|
Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [Inorganic ion transport and metabolism];
Pssm-ID: 443809 [Multi-domain] Cd Length: 229 Bit Score: 77.47 E-value: 2.73e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYR-----RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNI---- 1983
Cdd:COG4778 4 LLEVENLSKTFTlhlqgGKRLPVLDGVSFSVAAGECVALTGPSGAGKSTLLKCIYGNYLPDSGSILVRHDGGWVDLaqas 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1984 -HEVH----QNMGYCPQFdaiteL-----LTGREHVEFFALLRGVPEKE-VGKVGEWaIRKLGLvkyGEK----YASNYS 2048
Cdd:COG4778 84 pREILalrrRTIGYVSQF-----LrviprVSALDVVAEPLLERGVDREEaRARAREL-LARLNL---PERlwdlPPATFS 154
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2049 GGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRflwnCALSIVKE----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG4778 155 GGEQQRVNIARGFIADPPLLLLDEPTASLDAANRA----VVVELIEEakarGTAIIGIFHDEEVREAVADRVVDVTPFS 229
|
|
| NupO |
COG3845 |
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and ... |
900-1109 |
3.03e-15 |
|
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and metabolism];
Pssm-ID: 443055 [Multi-domain] Cd Length: 504 Bit Score: 81.23 E-value: 3.03e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVkVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI---------RSEMSS 970
Cdd:COG3845 259 EVENLS-VRDDRGVPALKDVSLEVRAGEILGIAGVAGNGQSELAEALAGLRPPASGSIRLDGEDItglsprerrRLGVAY 337
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 I---RQNLGVCPqhnvlfDMlTVEEHIWF-------YARLKGLSEKHVKAEMEQM--ALDVgLPPSkLKSKTSQLSGGMQ 1038
Cdd:COG3845 338 IpedRLGRGLVP------DM-SVAENLILgryrrppFSRGGFLDRKAIRAFAEELieEFDV-RTPG-PDTPARSLSGGNQ 408
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1039 RKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG3845 409 QKVILARELSRDPKLLIAAQPTRGLDVGAIEFIHQRLLELRdAGAAVLLISEDLDEILALSDRIAVMYEGRI 480
|
|
| YbbA |
COG4181 |
Predicted ABC-type transport system involved in lysophospholipase L1 biosynthesis, ATPase ... |
899-1109 |
3.50e-15 |
|
Predicted ABC-type transport system involved in lysophospholipase L1 biosynthesis, ATPase component [Secondary metabolites biosynthesis, transport and catabolism];
Pssm-ID: 443338 [Multi-domain] Cd Length: 233 Bit Score: 77.09 E-value: 3.50e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVY--RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIR 972
Cdd:COG4181 9 IELRGLTKTVgtGAGELTILKGISLEVEAGESVAIVGASGSGKSTLLGLLAGLDRPTSGTVRLAGQDLfaldEDARARLR 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 -QNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEqmaLD-VGL-------PpsklksktSQLSGGMQRKLSV 1043
Cdd:COG4181 89 aRHVGFVFQSFQLLPTLTALENVMLPLELAGRRDARARARAL---LErVGLghrldhyP--------AQLSGGEQQRVAL 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHhmDEADIL-GDRIAIISHGKL 1109
Cdd:COG4181 158 ARAFATEPAILFADEPTGNLDAATGEQIIDLLfeLNRERGTTLVLVTH--DPALAArCDRVLRLRAGRL 224
|
|
| PRK13657 |
PRK13657 |
glucan ABC transporter ATP-binding protein/ permease; |
910-1109 |
3.91e-15 |
|
glucan ABC transporter ATP-binding protein/ permease;
Pssm-ID: 184214 [Multi-domain] Cd Length: 588 Bit Score: 81.16 E-value: 3.91e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 910 DGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMl 988
Cdd:PRK13657 345 DNSRQGVEDVSFEAKPGQTVAIVGPTGAGKSTLINLLQRVFDPQSGRILIDGTDIRTvTRASLRRNIAVVFQDAGLFNR- 423
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIwfyaRL----KGLSEKHVKAEMEQmALD-VGLPPSKLKS----KTSQLSGGMQRKLSVALAFVGGSKVVILDEP 1059
Cdd:PRK13657 424 SIEDNI----RVgrpdATDEEMRAAAERAQ-AHDfIERKPDGYDTvvgeRGRQLSGGERQRLAIARALLKDPPILILDEA 498
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1060 TAGVDPYSRRGIWELLLKYRQGRT--II---LSThhMDEAdilgDRIAIISHGKL 1109
Cdd:PRK13657 499 TSALDVETEAKVKAALDELMKGRTtfIIahrLST--VRNA----DRILVFDNGRV 547
|
|
| Uup |
COG0488 |
ATPase components of ABC transporters with duplicated ATPase domains [General function ... |
901-1109 |
4.87e-15 |
|
ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only];
Pssm-ID: 440254 [Multi-domain] Cd Length: 520 Bit Score: 80.49 E-value: 4.87e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAyILGKDIRsemssirqnLGVCPQ 980
Cdd:COG0488 1 LENLSKSF--GGRPLLDDVSLSINPGDRIGLVGRNGAGKSTLLKILAGELEPDSGEV-SIPKGLR---------IGYLPQ 68
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLFDMLTVEEHIW--------FYARLKGLSEKHVKAEMEQMALD------------------------VGLPPSKLKS 1028
Cdd:COG0488 69 EPPLDDDLTVLDTVLdgdaelraLEAELEELEAKLAEPDEDLERLAelqeefealggweaearaeeilsgLGFPEEDLDR 148
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1029 KTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRgiW--ELLLKYRqGrTIILSTHhmDEA--DILGDRIAII 1104
Cdd:COG0488 149 PVSELSGGWRRRVALARALLSEPDLLLLDEPTNHLDLESIE--WleEFLKNYP-G-TVLVVSH--DRYflDRVATRILEL 222
|
....*
gi 568925948 1105 SHGKL 1109
Cdd:COG0488 223 DRGKL 227
|
|
| PRK10619 |
PRK10619 |
histidine ABC transporter ATP-binding protein HisP; |
909-1109 |
5.51e-15 |
|
histidine ABC transporter ATP-binding protein HisP;
Pssm-ID: 182592 [Multi-domain] Cd Length: 257 Bit Score: 77.32 E-value: 5.51e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR--------------SEMSSIRQN 974
Cdd:PRK10619 14 RYGEHEVLKGVSLQANAGDVISIIGSSGSGKSTFLRCINFLEKPSEGSIVVNGQTINlvrdkdgqlkvadkNQLRLLRTR 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHNVLFDMLTVEEHIWFY-ARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKV 1053
Cdd:PRK10619 94 LTMVFQHFNLWSHMTVLENVMEApIQVLGLSKQEARERAVKYLAKVGIDERAQGKYPVHLSGGQQQRVSIARALAMEPEV 173
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK10619 174 LLFDEPTSALDPELVGEVLRIMQQLaEEGKTMVVVTHEMGFARHVSSHVIFLHQGKI 230
|
|
| nikD |
PRK10418 |
nickel transporter ATP-binding protein NikD; Provisional |
898-1117 |
7.05e-15 |
|
nickel transporter ATP-binding protein NikD; Provisional
Pssm-ID: 236688 [Multi-domain] Cd Length: 254 Bit Score: 76.66 E-value: 7.05e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 898 GVSIQNLVkVYRDGmkVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPP----TSGTAYILGK-----DIRSEM 968
Cdd:PRK10418 4 QIELRNIA-LQAAQ--PLVHGVSLTLQRGRVLALVGGSGSGKSLTCAAALGILPAgvrqTAGRVLLDGKpvapcALRGRK 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 -SSIRQNlgvcPQH--NVLFDMLTveehiwfYAR--LKGLSEKHVKAEMEQMALDVGL--PPSKLKSKTSQLSGGMQRKL 1041
Cdd:PRK10418 81 iATIMQN----PRSafNPLHTMHT-------HARetCLALGKPADDATLTAALEAVGLenAARVLKLYPFEMSGGMLQRM 149
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1042 SVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL--CCVGSSLF 1117
Cdd:PRK10418 150 MIALALLCEAPFIIADEPTTDLDVVAQARILDLLesIVQKRALGMLLVTHDMGVVARLADDVAVMSHGRIveQGDVETLF 229
|
|
| ABC_ModC_molybdenum_transporter |
cd03297 |
ATP-binding cassette domain of the molybdenum transport system; ModC is an ABC-type ... |
1940-2123 |
9.01e-15 |
|
ATP-binding cassette domain of the molybdenum transport system; ModC is an ABC-type transporter and the ATPase component of a molybdate transport system that also includes the periplasmic binding protein ModA and the membrane protein ModB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213264 [Multi-domain] Cd Length: 214 Bit Score: 75.41 E-value: 9.01e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1940 PGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN--------KNSILSniheVHQ-NMGYCPQFDAITELLTGREHVE 2010
Cdd:cd03297 22 NEEVTGIFGASGAGKSTLLRCIAGLEKPDGGTIVLNgtvlfdsrKKINLP----PQQrKIGLVFQQYALFPHLNVRENLA 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2011 FfALLRGVPEKEVGKVGEWAIRkLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCAL 2090
Cdd:cd03297 98 F-GLKRKRNREDRISVDELLDL-LGLDHLLNRYPAQLSGGEKQRVALARALAAQPELLLLDEPFSALDRALRLQLLPELK 175
|
170 180 190
....*....|....*....|....*....|....
gi 568925948 2091 SIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03297 176 QIKKNlNIPVIFVTHDLSEAEYLADRIVVMEDGR 209
|
|
| PRK15439 |
PRK15439 |
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional |
918-1109 |
9.52e-15 |
|
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional
Pssm-ID: 185336 [Multi-domain] Cd Length: 510 Bit Score: 79.71 E-value: 9.52e-15
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 918 GLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGV--CPQHNVLFDMLTVEEHIW 995
Cdd:PRK15439 29 GIDFTLHAGEVHALLGGNGAGKSTLMKIIAGIVPPDSGTLEIGGNPCARLTPAKAHQLGIylVPQEPLLFPNLSVKENIL 108
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 996 FYARLKGLSEKHVKAEMEQMA--LDVGLPPSKLKSKTSQLSGGMqRKLsvalafVGGSKVVILDEPTAGVDPYSR----R 1069
Cdd:PRK15439 109 FGLPKRQASMQKMKQLLAALGcqLDLDSSAGSLEVADRQIVEIL-RGL------MRDSRILILDEPTASLTPAETerlfS 181
|
170 180 190 200
....*....|....*....|....*....|....*....|
gi 568925948 1070 GIWELLlkyRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK15439 182 RIRELL---AQGVGIVFISHKLPEIRQLADRISVMRDGTI 218
|
|
| AbcC |
COG1135 |
ABC-type methionine transport system, ATPase component [Amino acid transport and metabolism]; |
1913-2123 |
1.18e-14 |
|
ABC-type methionine transport system, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 440750 [Multi-domain] Cd Length: 339 Bit Score: 77.81 E-value: 1.18e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDAFLNKNSI--LSN--IHE 1985
Cdd:COG1135 1 MIELENLSKTFPTKGGPvtALDDVSLTIEKGEIFGIIGYSGAGKSTLIRCINLlERP-TSGSVLVDGVDLtaLSEreLRA 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYCPQ-FDaiteLLTGR---EHVEfFAL-LRGVPEKEV-GKVGEwairkL----GLVKYGEKYASNYSGGNKRKL 2055
Cdd:COG1135 80 ARRKIGMIFQhFN----LLSSRtvaENVA-LPLeIAGVPKAEIrKRVAE-----LlelvGLSDKADAYPSQLSGGQKQRV 149
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2056 STAMALIGGPPVVFLDEPTTGMDPKARRflwncalSIV--------KEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG1135 150 GIARALANNPKVLLCDEATSALDPETTR-------SILdllkdinrELGLTIVLITHEMDVVRRICDRVAVLENGR 218
|
|
| cbiO |
PRK13643 |
energy-coupling factor transporter ATPase; |
1938-2108 |
1.31e-14 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184203 [Multi-domain] Cd Length: 288 Bit Score: 76.70 E-value: 1.31e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTG-----DTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQFDAiTELL--TGREHVE 2010
Cdd:PRK13643 29 VKKGSYTALIGHTGSGKSTLLQHLNGllqptEGKVTVGDIVVSSTSKQKEIKPVRKKVGVVFQFPE-SQLFeeTVLKDVA 107
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2011 FFALLRGVPEKEVGKVGEWAIRKLGLVK-YGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCA 2089
Cdd:PRK13643 108 FGPQNFGIPKEKAEKIAAEKLEMVGLADeFWEKSPFELSGGQMRRVAIAGILAMEPEVLVLDEPTAGLDPKARIEMMQLF 187
|
170
....*....|....*....
gi 568925948 2090 LSIVKEGRSVVLTSHSMEE 2108
Cdd:PRK13643 188 ESIHQSGQTVVLVTHLMDD 206
|
|
| PRK11264 |
PRK11264 |
putative amino-acid ABC transporter ATP-binding protein YecC; Provisional |
899-1109 |
1.32e-14 |
|
putative amino-acid ABC transporter ATP-binding protein YecC; Provisional
Pssm-ID: 183063 [Multi-domain] Cd Length: 250 Bit Score: 75.94 E-value: 1.32e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYI------LGKDIRSEMSSIR 972
Cdd:PRK11264 4 IEVKNLVKKFHG--QTVLHGIDLEVKPGEVVAIIGPSGSGKTTLLRCINLLEQPEAGTIRVgditidTARSLSQQKGLIR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 ---QNLGVCPQHNVLFDMLTVEEHIwfyarLKG--LSEKHVKAEMEQMALD----VGLPpSKLKSKTSQLSGGMQRKLSV 1043
Cdd:PRK11264 82 qlrQHVGFVFQNFNLFPHRTVLENI-----IEGpvIVKGEPKEEATARAREllakVGLA-GKETSYPRRLSGGQQQRVAI 155
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPysrRGIWELLLKYRQ----GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK11264 156 ARALAMRPEVILFDEPTSALDP---ELVGEVLNTIRQlaqeKRTMVIVTHEMSFARDVADRAIFMDQGRI 222
|
|
| cbiO |
PRK13650 |
energy-coupling factor transporter ATPase; |
1912-2182 |
1.59e-14 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184209 [Multi-domain] Cd Length: 279 Bit Score: 76.31 E-value: 1.59e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1912 DILEIKELTKIYRR-KRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFL--------NKNSILSN 1982
Cdd:PRK13650 3 NIIEVKNLTFKYKEdQEKYTLNDVSFHVKQGEWLSIIGHNGSGKSTTVRLIDGLLEAESGQIIIdgdllteeNVWDIRHK 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1983 IHEVHQNmgycP--QFDAITElltgREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:PRK13650 83 IGMVFQN----PdnQFVGATV----EDDVAFGLENKGIPHEEMKERVNEALELVGMQDFKEREPARLSGGQKQRVAIAGA 154
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECeALCTRMAIMVNGRFRCLGSVQHLKNRFGD 2139
Cdd:PRK13650 155 VAMRPKIIILDEATSMLDPEGRLELIKTIKGIRDDyQMTVISITHDLDEV-ALSDRVLVMKNGQVESTSTPRELFSRGND 233
|
250 260 270 280
....*....|....*....|....*....|....*....|...
gi 568925948 2140 GYTIVVRIAGSNPDLKPVQEfFGLAFPGSVLKEKHrnmLQYQL 2182
Cdd:PRK13650 234 LLQLGLDIPFTTSLVQSLRQ-NGYDLPEGYLTEKE---LEEQL 272
|
|
| MglA |
COG1129 |
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism]; |
1913-2123 |
1.61e-14 |
|
ABC-type sugar transport system, ATPase component [Carbohydrate transport and metabolism];
Pssm-ID: 440745 [Multi-domain] Cd Length: 497 Bit Score: 78.91 E-value: 1.61e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTkiyrrkRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHE-VHQNM 1990
Cdd:COG1129 256 VLEVEGLS------VGGVVRDVSFSVRAGEILGIAGLVGAGRTELARALFGADPADSGEIRLDGKPVrIRSPRDaIRAGI 329
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQ-------------FDAITelLTGREHVEFFALLRgvpEKEVGKVGEWAIRKLGlVKYG--EKYASNYSGGNKRKL 2055
Cdd:COG1129 330 AYVPEdrkgeglvldlsiRENIT--LASLDRLSRGGLLD---RRRERALAEEYIKRLR-IKTPspEQPVGNLSGGNQQKV 403
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2056 STAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG1129 404 VLAKWLATDPKVLILDEPTRGIDVGAKAEIYRLIRELAAEGKAVIVISSELPELLGLSDRILVMREGR 471
|
|
| PRK11831 |
PRK11831 |
phospholipid ABC transporter ATP-binding protein MlaF; |
903-1120 |
1.66e-14 |
|
phospholipid ABC transporter ATP-binding protein MlaF;
Pssm-ID: 236997 [Multi-domain] Cd Length: 269 Bit Score: 75.96 E-value: 1.66e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 903 NLVKV----YRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQN 974
Cdd:PRK11831 6 NLVDMrgvsFTRGNRCIFDNISLTVPRGKITAIMGPSGIGKTTLLRLIGGQIAPDHGEILFDGENIpamsRSRLYTVRKR 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHNVLFDMLTVEEHIWFYARLKG-LSEKHVKAEMeQMALD-VGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:PRK11831 86 MSMLFQSGALFTDMNVFDNVAYPLREHTqLPAPLLHSTV-MMKLEaVGLRGAA-KLMPSELSGGMARRAALARAIALEPD 163
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFLKN 1120
Cdd:PRK11831 164 LIMFDEPFVGQDPITMGVLVKLIseLNSALGVTCVVVSHDVPEVLSIADHAYIVADKKIVAHGSAQALQA 233
|
|
| PRK09700 |
PRK09700 |
D-allose ABC transporter ATP-binding protein AlsA; |
901-1109 |
1.67e-14 |
|
D-allose ABC transporter ATP-binding protein AlsA;
Pssm-ID: 182036 [Multi-domain] Cd Length: 510 Bit Score: 78.67 E-value: 1.67e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKvyRDGMKVAVDGLALNfyEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI--RSEMSSIRQNLGVC 978
Cdd:PRK09700 268 VRNVTS--RDRKKVRDISFSVC--RGEILGFAGLVGSGRTELMNCLFGVDKRAGGEIRLNGKDIspRSPLDAVKKGMAYI 343
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 PQH---NVLFDMLTVEEHIWFY-----ARLKG----LSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALA 1046
Cdd:PRK09700 344 TESrrdNGFFPNFSIAQNMAISrslkdGGYKGamglFHEVDEQRTAENQRELLALKCHSVNQNITELSGGNQQKVLISKW 423
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1047 FVGGSKVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK09700 424 LCCCPEVIIFDEPTRGIDVGAKAEIYKVMRQLaDDGKVILMVSSELPEIITVCDRIAVFCEGRL 487
|
|
| 3a01208 |
TIGR00958 |
Conjugate Transporter-2 (CT2) Family protein; [Transport and binding proteins, Other] |
918-1131 |
1.83e-14 |
|
Conjugate Transporter-2 (CT2) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273363 [Multi-domain] Cd Length: 711 Bit Score: 79.38 E-value: 1.83e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 918 GLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMlTVEEHIwF 996
Cdd:TIGR00958 499 GLTFTLHPGEVVALVGPSGSGKSTVAALLQNLYQPTGGQVLLDGVPLVQyDHHYLHRQVALVGQEPVLFSG-SVRENI-A 576
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 997 YARLKGLSEKHVKAEMEQMALD--VGLPP---SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGI 1071
Cdd:TIGR00958 577 YGLTDTPDEEIMAAAKAANAHDfiMEFPNgydTEVGEKGSQLSGGQKQRIAIARALVRKPRVLILDEATSALDAECEQLL 656
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1072 WEllLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGSSLFLKNQlgTGYYLTLV 1131
Cdd:TIGR00958 657 QE--SRSRASRTVLLIAHRLSTVE-RADQILVLKKGSVVEMGTHKQLMED--QGCYKHLV 711
|
|
| ABC_HisP_GlnQ |
cd03262 |
ATP-binding cassette domain of the histidine and glutamine transporters; HisP and GlnQ are the ... |
1914-2123 |
2.92e-14 |
|
ATP-binding cassette domain of the histidine and glutamine transporters; HisP and GlnQ are the ATP-binding components of the bacterial periplasmic histidine and glutamine permeases, respectively. Histidine permease is a multi-subunit complex containing the HisQ and HisM integral membrane subunits and two copies of HisP. HisP has properties intermediate between those of integral and peripheral membrane proteins and is accessible from both sides of the membrane, presumably by its interaction with HisQ and HisM. The two HisP subunits form a homodimer within the complex. The domain structure of the amino acid uptake systems is typical for prokaryotic extracellular solute binding protein-dependent uptake systems. All of the amino acid uptake systems also have at least one, and in a few cases, two extracellular solute binding proteins located in the periplasm of Gram-negative bacteria, or attached to the cell membrane of Gram-positive bacteria. The best-studied member of the PAAT (polar amino acid transport) family is the HisJQMP system of S. typhimurium, where HisJ is the extracellular solute binding proteins and HisP is the ABC protein.
Pssm-ID: 213229 [Multi-domain] Cd Length: 213 Bit Score: 74.10 E-value: 2.92e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS---NIHEVHQNM 1990
Cdd:cd03262 1 IEIKNLHKSF--GDFHVLKGIDLTVKKGEVVVIIGPSGSGKSTLLRCINLLEEPDSGTIIIDGLKLTDdkkNINELRQKV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQ----------FDAITELLTgrehveffaLLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:cd03262 79 GMVFQqfnlfphltvLENITLAPI---------KVKGMSKAEAEERALELLEKVGLADKADAYPAQLSGGQQQRVAIARA 149
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03262 150 LAMNPKVMLFDEPTSALDPELVGEVLDVMKDLAEEGMTMVVVTHEMGFAREVADRVIFMDDGR 212
|
|
| TauB |
COG4525 |
ABC-type taurine transport system, ATPase component [Inorganic ion transport and metabolism]; |
1914-2119 |
3.23e-14 |
|
ABC-type taurine transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443596 [Multi-domain] Cd Length: 262 Bit Score: 74.90 E-value: 3.23e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIY--RRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHE---VHQ 1988
Cdd:COG4525 4 LTVRHVSVRYpgGGQPQPALQDVSLTIESGEFVVALGASGCGKTTLLNLIAGFLAPSSGEITLDGVPVTGPGADrgvVFQ 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NmgycpqfDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:COG4525 84 K-------DALLPWLNVLDNVAFGLRLRGVPKAERRARAEELLALVGLADFARRRIWQLSGGMRQRVGIARALAADPRFL 156
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIM 2119
Cdd:COG4525 157 LMDEPFGALDALTREQMQELLLDVWQRtGKGVFLITHSVEEALFLATRLVVM 208
|
|
| ABCC_cytochrome_bd |
cd03247 |
ATP-binding cassette domain of CydCD, subfamily C; The CYD subfamily implicated in cytochrome ... |
1914-2124 |
3.39e-14 |
|
ATP-binding cassette domain of CydCD, subfamily C; The CYD subfamily implicated in cytochrome bd biogenesis. The CydC and CydD proteins are important for the formation of cytochrome bd terminal oxidase of E. coli and it has been proposed that they were necessary for biosynthesis of the cytochrome bd quinol oxidase and for periplasmic c-type cytochromes. CydCD were proposed to determine a heterooligomeric complex important for heme export into the periplasm or to be involved in the maintenance of the proper redox state of the periplasmic space. In Bacillus subtilis, the absence of CydCD does not affect the presence of halo-cytochrome c in the membrane and this observation suggests that CydCD proteins are not involved in the export of heme in this organism.
Pssm-ID: 213214 [Multi-domain] Cd Length: 178 Bit Score: 72.73 E-value: 3.39e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:cd03247 1 LSINNVSFSYPEQEQQVLKNLSLELKQGEKIALLGRSGSGKSTLLQLLTGDLKPQQGEITLDGVPVSDLEKALSSLISVL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQfdaitelltgREHVeFFALLRgvpeKEVGKvgewairklglvkygekyasNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03247 81 NQ----------RPYL-FDTTLR----NNLGR--------------------RFSGGERQRLALARILLQDAPIVLLDEP 125
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSiVKEGRSVVLTSHSMEECEALcTRMAIMVNGRF 2124
Cdd:cd03247 126 TVGLDPITERQLLSLIFE-VLKDKTLIWITHHLTGIEHM-DKILFLENGKI 174
|
|
| cbiO |
PRK13635 |
energy-coupling factor ABC transporter ATP-binding protein; |
1910-2123 |
5.61e-14 |
|
energy-coupling factor ABC transporter ATP-binding protein;
Pssm-ID: 184195 [Multi-domain] Cd Length: 279 Bit Score: 74.67 E-value: 5.61e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-----DTPVTRGDAFLNKNS---ILS 1981
Cdd:PRK13635 2 KEEIIRVEHISFRYPDAATYALKDVSFSVYEGEWVAIVGHNGSGKSTLAKLLNGlllpeAGTITVGGMVLSEETvwdVRR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1982 NIHEVHQNmgycP--QFDAITElltgREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAM 2059
Cdd:PRK13635 82 QVGMVFQN----PdnQFVGATV----QDDVAFGLENIGVPREEMVERVDQALRQVGMEDFLNREPHRLSGGQKQRVAIAG 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2060 ALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLT-SHSMEECeALCTRMAIMVNGR 2123
Cdd:PRK13635 154 VLALQPDIIILDEATSMLDPRGRREVLETVRQLKEQKGITVLSiTHDLDEA-AQADRVIVMNKGE 217
|
|
| thiQ |
PRK10771 |
thiamine ABC transporter ATP-binding protein ThiQ; |
932-1109 |
5.66e-14 |
|
thiamine ABC transporter ATP-binding protein ThiQ;
Pssm-ID: 182716 [Multi-domain] Cd Length: 232 Bit Score: 73.46 E-value: 5.66e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 932 LGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSeMSSIRQNLGVCPQHNVLFDMLTVEEHIWF--YARLKgLSEKHvK 1009
Cdd:PRK10771 31 LGPSGAGKSTLLNLIAGFLTPASGSLTLNGQDHTT-TPPSRRPVSMLFQENNLFSHLTVAQNIGLglNPGLK-LNAAQ-R 107
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1010 AEMEQMALDVGLpPSKLKSKTSQLSGGmQRKlSVALA--FVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGR--TII 1085
Cdd:PRK10771 108 EKLHAIARQMGI-EDLLARLPGQLSGG-QRQ-RVALArcLVREQPILLLDEPFSALDPALRQEMLTLVSQVCQERqlTLL 184
|
170 180
....*....|....*....|....
gi 568925948 1086 LSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK10771 185 MVSHSLEDAARIAPRSLVVADGRI 208
|
|
| PRK09984 |
PRK09984 |
phosphonate ABC transporter ATP-binding protein; |
1913-2147 |
6.40e-14 |
|
phosphonate ABC transporter ATP-binding protein;
Pssm-ID: 182182 [Multi-domain] Cd Length: 262 Bit Score: 74.28 E-value: 6.40e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFK----MLTGD-TPVTR----GDAFLNKNSILSNI 1983
Cdd:PRK09984 4 IIRVEKLAKTFNQHQ--ALHAVDLNIHHGEMVALLGPSGSGKSTLLRhlsgLITGDkSAGSHiellGRTVQREGRLARDI 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1984 HEVHQNMGYCPQFDAITELLTGREHVEFFAL---------LRGVPEKEVGKVGEwAIRKLGLVKYGEKYASNYSGGNKRK 2054
Cdd:PRK09984 82 RKSRANTGYIFQQFNLVNRLSVLENVLIGALgstpfwrtcFSWFTREQKQRALQ-ALTRVGMVHFAHQRVSTLSGGQQQR 160
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2055 LSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVK-EGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK09984 161 VAIARALMQQAKVILADEPIASLDPESARIVMDTLRDINQnDGITVVVTLHQVDYALRYCERIVALRQGHVFYDGSSQQF 240
|
250
....*....|....*
gi 568925948 2134 KN-RFGDGYTIVVRI 2147
Cdd:PRK09984 241 DNeRFDHLYRSINRV 255
|
|
| fecE |
PRK11231 |
Fe(3+) dicitrate ABC transporter ATP-binding protein FecE; |
1913-2123 |
6.84e-14 |
|
Fe(3+) dicitrate ABC transporter ATP-binding protein FecE;
Pssm-ID: 183044 [Multi-domain] Cd Length: 255 Bit Score: 73.89 E-value: 6.84e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPVTrGDAFLNKNSILS-NIHEVHQNM 1990
Cdd:PRK11231 2 TLRTENLTVGYGTKR--ILNDLSLSLPTGKITALIGPNGCGKSTLLKCFARlLTPQS-GTVFLGDKPISMlSSRQLARRL 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQFDAITELLTGREHVE--------FFALLRGVPEKEVgkvgEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALI 2062
Cdd:PRK11231 79 ALLPQHHLTPEGITVRELVAygrspwlsLWGRLSAEDNARV----NQAMEQTRINHLADRRLTDLSGGQRQRAFLAMVLA 154
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2063 GGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK11231 155 QDTPVVLLDEPTTYLDINHQVELMRLMRELNTQGKTVVTVLHDLNQASRYCDHLVVLANGH 215
|
|
| btuD |
PRK09536 |
corrinoid ABC transporter ATPase; Reviewed |
899-1113 |
7.54e-14 |
|
corrinoid ABC transporter ATPase; Reviewed
Pssm-ID: 236554 [Multi-domain] Cd Length: 402 Bit Score: 76.03 E-value: 7.54e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVkVYRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMS-SIRQNLGV 977
Cdd:PRK09536 4 IDVSDLS-VEFGDTTV-LDGVDLSVREGSLVGLVGPNGAGKTTLLRAINGTLTPTAGTVLVAGDDVEALSArAASRRVAS 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVL---FDMLTVEE-----HIWFYARLKGLSEKHVKAEMEQmaldVGLPPSKLKSKTSqLSGGMQRKLSVALAFVG 1049
Cdd:PRK09536 82 VPQDTSLsfeFDVRQVVEmgrtpHRSRFDTWTETDRAAVERAMER----TGVAQFADRPVTS-LSGGERQRVLLARALAQ 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1050 GSKVVILDEPTAGVDpySRRGIWELLLKYR---QGRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:PRK09536 157 ATPVLLLDEPTASLD--INHQVRTLELVRRlvdDGKTAVAAIHDLDLAARYCDELVLLADGRVRAAG 221
|
|
| PRK13538 |
PRK13538 |
cytochrome c biogenesis heme-transporting ATPase CcmA; |
1914-2104 |
8.22e-14 |
|
cytochrome c biogenesis heme-transporting ATPase CcmA;
Pssm-ID: 184125 [Multi-domain] Cd Length: 204 Bit Score: 72.53 E-value: 8.22e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIyrRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYC 1993
Cdd:PRK13538 2 LEARNLACE--RDERILFSGLSFTLNAGELVQIEGPNGAGKTSLLRILAGLARPDAGEVLWQGEPIRRQRDEYHQDLLYL 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFALLRGVPEKEVgkvgEW-AIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDE 2072
Cdd:PRK13538 80 GHQPGIKTELTALENLRFYQRLHGPGDDEA----LWeALAQVGLAGFEDVPVRQLSAGQQRRVALARLWLTRAPLWILDE 155
|
170 180 190
....*....|....*....|....*....|....*..
gi 568925948 2073 PTTGMDPKA-----RRFLWNCAlsivkEGRSVVLTSH 2104
Cdd:PRK13538 156 PFTAIDKQGvarleALLAQHAE-----QGGMVILTTH 187
|
|
| tauB |
PRK11248 |
taurine ABC transporter ATP-binding subunit; |
1928-2142 |
8.46e-14 |
|
taurine ABC transporter ATP-binding subunit;
Pssm-ID: 183056 [Multi-domain] Cd Length: 255 Bit Score: 73.58 E-value: 8.46e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHE---VHQNMGYCPQFDAItellt 2004
Cdd:PRK11248 14 KPALEDINLTLESGELLVVLGPSGCGKTTLLNLIAGFVPYQHGSITLDGKPVEGPGAErgvVFQNEGLLPWRNVQ----- 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2005 grEHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRF 2084
Cdd:PRK11248 89 --DNVAFGLQLAGVEKMQRLEIAHQMLKKVGLEGAEKRYIWQLSGGQRQRVGIARALAANPQLLLLDEPFGALDAFTREQ 166
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2085 LWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRClgsVQHLKNRFGDGYT 2142
Cdd:PRK11248 167 MQTLLLKLWQEtGKQVLLITHDIEEAVFMATELVLLSPGPGRV---VERLPLNFARRFV 222
|
|
| cbiO |
PRK13648 |
cobalt transporter ATP-binding subunit; Provisional |
915-1109 |
8.99e-14 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184207 [Multi-domain] Cd Length: 269 Bit Score: 74.02 E-value: 8.99e-14
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGVCPQH-NVLFDMLTVEE 992
Cdd:PRK13648 24 TLKDVSFNIPKGQWTSIVGHNGSGKSTIAKLMIGIEKVKSGEIFYNNQAITDDnFEKLRKHIGIVFQNpDNQFVGSIVKY 103
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 993 HIWFYARLKGLSEKHVKAEMEQMALDVGLpPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIW 1072
Cdd:PRK13648 104 DVAFGLENHAVPYDEMHRRVSEALKQVDM-LERADYEPNALSGGQKQRVAIAGVLALNPSVIILDEATSMLDPDARQNLL 182
|
170 180 190
....*....|....*....|....*....|....*....
gi 568925948 1073 ELLLKYRQGR--TIILSTHHMDEAdILGDRIAIISHGKL 1109
Cdd:PRK13648 183 DLVRKVKSEHniTIISITHDLSEA-MEADHVIVMNKGTV 220
|
|
| ABCC_Protease_Secretion |
cd03246 |
ATP-binding cassette domain of PrtD, subfamily C; This family represents the ABC component of ... |
1914-2123 |
1.17e-13 |
|
ATP-binding cassette domain of PrtD, subfamily C; This family represents the ABC component of the protease secretion system PrtD, a 60-kDa integral membrane protein sharing 37% identity with HlyB, the ABC component of the alpha-hemolysin secretion pathway, in the C-terminal domain. They export degradative enzymes by using a type I protein secretion system and lack an N-terminal signal peptide, but contain a C-terminal secretion signal. The Type I secretion apparatus is made up of three components, an ABC transporter, a membrane fusion protein (MFP), and an outer membrane protein (OMP). For the HlyA transporter complex, HlyB (ABC transporter) and HlyD (MFP) reside in the inner membrane of E. coli. The OMP component is TolC, which is thought to interact with the MFP to form a continuous channel across the periplasm from the cytoplasm to the exterior. HlyB belongs to the family of ABC transporters, which are ubiquitous, ATP-dependent transmembrane pumps or channels. The spectrum of transport substrates ranges from inorganic ions, nutrients such as amino acids, sugars, or peptides, hydrophobic drugs, to large polypeptides, such as HlyA.
Pssm-ID: 213213 [Multi-domain] Cd Length: 173 Bit Score: 71.09 E-value: 1.17e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNkNSILSNIHEVH--QNMG 1991
Cdd:cd03246 1 LEVENVSFRYPGAEPPVLRNVSFSIEPGESLAIIGPSGSGKSTLARLILGLLRPTSGRVRLD-GADISQWDPNElgDHVG 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDaitELLTGrehveffallrgvpekevgkvgewAIrklglvkygekyASN-YSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:cd03246 80 YLPQDD---ELFSG------------------------SI------------AENiLSGGQRQRLGLARALYGNPRILVL 120
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEAlCTRMAIMVNGR 2123
Cdd:cd03246 121 DEPNSHLDVEGERALNQAIAALKAAGATRIVIAHRPETLAS-ADRILVLEDGR 172
|
|
| nikE |
PRK10419 |
nickel ABC transporter ATP-binding protein NikE; |
900-1109 |
1.37e-13 |
|
nickel ABC transporter ATP-binding protein NikE;
Pssm-ID: 236689 [Multi-domain] Cd Length: 268 Bit Score: 73.18 E-value: 1.37e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDG-------MKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEM 968
Cdd:PRK10419 5 NVSGLSHHYAHGglsgkhqHQTVLNNVSLSLKSGETVALLGRSGCGKSTLARLLVGLESPSQGNVSWRGEPLaklnRAQR 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 SSIRQNL---------GVCPQHnvlfdmlTVEEHIWFYAR-LKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQ 1038
Cdd:PRK10419 85 KAFRRDIqmvfqdsisAVNPRK-------TVREIIREPLRhLLSLDKAERLARASEMLRAVDLDDSVLDKRPPQLSGGQL 157
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1039 RKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK10419 158 QRVCLARALAVEPKLLILDEAVSNLDLVLQAGVIRLLKKLQQqfGTACLFITHDLRLVERFCQRVMVMDNGQI 230
|
|
| PRK10247 |
PRK10247 |
putative ABC transporter ATP-binding protein YbbL; Provisional |
908-1107 |
1.94e-13 |
|
putative ABC transporter ATP-binding protein YbbL; Provisional
Pssm-ID: 182331 [Multi-domain] Cd Length: 225 Bit Score: 72.05 E-value: 1.94e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMS--SIRQNLGVCPQHNVLF 985
Cdd:PRK10247 15 YLAGDAKILNNISFSLRAGEFKLITGPSGCGKSTLLKIVASLISPTSGTLLFEGEDI-STLKpeIYRQQVSYCAQTPTLF 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 DMlTVEEHIWF--YARLKGLSEKHVKAEMEQMaldvGLPPSKLKSKTSQLSGGMQRKLSVA--LAFVggSKVVILDEPTA 1061
Cdd:PRK10247 94 GD-TVYDNLIFpwQIRNQQPDPAIFLDDLERF----ALPDTILTKNIAELSGGEKQRISLIrnLQFM--PKVLLLDEITS 166
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1062 GVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHG 1107
Cdd:PRK10247 167 ALDESNKHNVNEIIHRYvrEQNIAVLWVTHDKDEINHADKVITLQPHA 214
|
|
| ntrCD |
TIGR01184 |
nitrate transport ATP-binding subunits C and D; This model describes the ATP binding subunits ... |
1931-2122 |
2.21e-13 |
|
nitrate transport ATP-binding subunits C and D; This model describes the ATP binding subunits of nitrate transport in bacteria and archaea. This protein belongs to the ATP-binding cassette (ABC) superfamily. It is thought that the two subunits encoded by ntrC and ntrD form the binding surface for interaction with ATP. This model is restricted in identifying ATP binding subunit associated with the nitrate transport. Nitrate assimilation is aided by other proteins derived from the operon which among others include products of ntrA - a regulatory protein; ntrB - a hydropbobic transmembrane permease and narB - a reductase. [Transport and binding proteins, Anions, Transport and binding proteins, Other]
Pssm-ID: 130252 [Multi-domain] Cd Length: 230 Bit Score: 71.73 E-value: 2.21e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1931 VDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHE---VHQNMGYCPqfdaiteLLTGRE 2007
Cdd:TIGR01184 1 LKGVNLTIQQGEFISLIGHSGCGKSTLLNLISGLAQPTSGGVILEGKQITEPGPDrmvVFQNYSLLP-------WLTVRE 73
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 HVEFF--ALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFL 2085
Cdd:TIGR01184 74 NIALAvdRVLPDLSKSERRAIVEEHIALVGLTEAADKRPGQLSGGMKQRVAIARALSIRPKVLLLDEPFGALDALTRGNL 153
|
170 180 190
....*....|....*....|....*....|....*...
gi 568925948 2086 WNCALSIVKEGR-SVVLTSHSMEECEALCTRMAIMVNG 2122
Cdd:TIGR01184 154 QEELMQIWEEHRvTVLMVTHDVDEALLLSDRVVMLTNG 191
|
|
| PRK14258 |
PRK14258 |
phosphate ABC transporter ATP-binding protein; Provisional |
898-1102 |
2.25e-13 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 184593 [Multi-domain] Cd Length: 261 Bit Score: 72.38 E-value: 2.25e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 898 GVSIQNLvKVYRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTS-----GTAYILGKDI---RSEMS 969
Cdd:PRK14258 7 AIKVNNL-SFYYDTQKI-LEGVSMEIYQSKVTAIIGPSGCGKSTFLKCLNRMNELESevrveGRVEFFNQNIyerRVNLN 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 970 SIRQNLG-VCPQHNvLFDMlTVEEHIWFYARLKGLSEK-HVKAEMEQMALDVGL---PPSKLKSKTSQLSGGMQRKLSVA 1044
Cdd:PRK14258 85 RLRRQVSmVHPKPN-LFPM-SVYDNVAYGVKIVGWRPKlEIDDIVESALKDADLwdeIKHKIHKSALDLSGGQQQRLCIA 162
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIA 1102
Cdd:PRK14258 163 RALAVKPKVLLMDEPCFGLDPIASMKVESLIqsLRLRSELTMVIVSHNLHQVSRLSDFTA 222
|
|
| 3a01204 |
TIGR00955 |
The Eye Pigment Precursor Transporter (EPP) Family protein; [Transport and binding proteins, ... |
1924-2143 |
2.49e-13 |
|
The Eye Pigment Precursor Transporter (EPP) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273361 [Multi-domain] Cd Length: 617 Bit Score: 75.47 E-value: 2.49e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1924 RRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP--VTR-GDAFLNKNSIlsNIHEVHQNMGYCPQFDAIT 2000
Cdd:TIGR00955 34 ERPRKHLLKNVSGVAKPGELLAVMGSSGAGKTTLMNALAFRSPkgVKGsGSVLLNGMPI--DAKEMRAISAYVQQDDLFI 111
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 ELLTGREHVEFFALLR---GVPEKEVGKVGEWAIRKLGLVKYG------EKYASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:TIGR00955 112 PTLTVREHLMFQAHLRmprRVTKKEKRERVDEVLQALGLRKCAntrigvPGRVKGLSGGERKRLAFASELLTDPPLLFCD 191
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2072 EPTTGMDPkarrFLWNCALSIVKE----GRSVVLTSH--SMEECEaLCTRMAIMVNGRFRCLGSVQHLKNRFGD-GYTI 2143
Cdd:TIGR00955 192 EPTSGLDS----FMAYSVVQVLKGlaqkGKTIICTIHqpSSELFE-LFDKIILMAEGRVAYLGSPDQAVPFFSDlGHPC 265
|
|
| PRK09984 |
PRK09984 |
phosphonate ABC transporter ATP-binding protein; |
899-1121 |
2.62e-13 |
|
phosphonate ABC transporter ATP-binding protein;
Pssm-ID: 182182 [Multi-domain] Cd Length: 262 Bit Score: 72.35 E-value: 2.62e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMkvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP----PTSGTAYI---------LGKDIR 965
Cdd:PRK09984 5 IRVEKLAKTFNQHQ--ALHAVDLNIHHGEMVALLGPSGSGKSTLLRHLSGLITgdksAGSHIELLgrtvqregrLARDIR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 966 SEmssiRQNLGVCPQHNVLFDMLTVEEHIW--------FYARLKGLSEKHVKAEMEQMALDVGLPPSKlKSKTSQLSGGM 1037
Cdd:PRK09984 83 KS----RANTGYIFQQFNLVNRLSVLENVLigalgstpFWRTCFSWFTREQKQRALQALTRVGMVHFA-HQRVSTLSGGQ 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1038 QRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVGSS 1115
Cdd:PRK09984 158 QQRVAIARALMQQAKVILADEPIASLDPESARIVMDTLRDINQndGITVVVTLHQVDYALRYCERIVALRQGHVFYDGSS 237
|
....*.
gi 568925948 1116 LFLKNQ 1121
Cdd:PRK09984 238 QQFDNE 243
|
|
| xylG |
TIGR02633 |
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose ... |
899-1108 |
2.69e-13 |
|
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose isomerase and xylulokinase enzymes for xylose utilization. Members of this protein family are the ATP-binding cassette (ABC) subunit of the known or predicted high-affinity xylose ABC transporter for xylose import. These genes, which closely resemble other sugar transport ABC transporter genes, typically are encoded near xylose utilization enzymes and regulatory proteins. Note that this form of the transporter contains two copies of the ABC transporter domain (pfam00005). [Transport and binding proteins, Carbohydrates, organic alcohols, and acids]
Pssm-ID: 131681 [Multi-domain] Cd Length: 500 Bit Score: 74.86 E-value: 2.69e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP--PTSGTAYILGKDIRSemSSIRQN-- 974
Cdd:TIGR02633 2 LEMKGIVKTF-GGVK-ALDGIDLEVRPGECVGLCGENGAGKSTLMKILSGVYPhgTWDGEIYWSGSPLKA--SNIRDTer 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 --LGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEM----EQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:TIGR02633 78 agIVIIHQELTLVPELSVAENIFLGNEITLPGGRMAYNAMylraKNLLRELQLDADNVTRPVGDYGGGQQQLVEIAKALN 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELL--LKyRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:TIGR02633 158 KQARLLILDEPSSSLTEKETEILLDIIrdLK-AHGVACVYISHKLNEVKAVCDTICVIRDGQ 218
|
|
| PLN03211 |
PLN03211 |
ABC transporter G-25; Provisional |
1938-2104 |
5.81e-13 |
|
ABC transporter G-25; Provisional
Pssm-ID: 215634 [Multi-domain] Cd Length: 659 Bit Score: 74.53 E-value: 5.81e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTpvtRGDAFLNKnsILSN----IHEVHQNMGYCPQFDAITELLTGREHVEFFA 2013
Cdd:PLN03211 91 ASPGEILAVLGPSGSGKSTLLNALAGRI---QGNNFTGT--ILANnrkpTKQILKRTGFVTQDDILYPHLTVRETLVFCS 165
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2014 LLR---GVPEKEVGKVGEWAIRKLGLVK-----YGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFL 2085
Cdd:PLN03211 166 LLRlpkSLTKQEKILVAESVISELGLTKcentiIGNSFIRGISGGERKRVSIAHEMLINPSLLILDEPTSGLDATAAYRL 245
|
170
....*....|....*....
gi 568925948 2086 WNCALSIVKEGRSVVLTSH 2104
Cdd:PLN03211 246 VLTLGSLAQKGKTIVTSMH 264
|
|
| PRK14271 |
PRK14271 |
phosphate ABC transporter ATP-binding protein; Provisional |
913-1109 |
6.42e-13 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172759 [Multi-domain] Cd Length: 276 Bit Score: 71.67 E-value: 6.42e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 913 KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAY-----ILGKDIRS--EMSSIRQNLGVCPQHNVLF 985
Cdd:PRK14271 34 KTVLDQVSMGFPARAVTSLMGPTGSGKTTFLRTLNRMNDKVSGYRYsgdvlLGGRSIFNyrDVLEFRRRVGMLFQRPNPF 113
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 DMLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGL---PPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:PRK14271 114 PMSIMDNVLAGVRAHKLVPRKEFRGVAQARLTEVGLwdaVKDRLSDSPFRLSGGQQQLLCLARTLAVNPEVLLLDEPTSA 193
|
170 180 190 200
....*....|....*....|....*....|....*....|....*..
gi 568925948 1063 VDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK14271 194 LDPTTTEKIEEFIRSLADRLTVIIVTHNLAQAARISDRAALFFDGRL 240
|
|
| ABCC_Glucan_exporter_like |
cd03254 |
ATP-binding cassette domain of glucan transporter and related proteins, subfamily C; Glucan ... |
1915-2104 |
7.29e-13 |
|
ATP-binding cassette domain of glucan transporter and related proteins, subfamily C; Glucan exporter ATP-binding protein. In A. tumefaciens cyclic beta-1, 2-glucan must be transported into the periplasmic space to exert its action as a virulence factor. This subfamily belongs to the MRP-like family and is involved in drug, peptide, and lipid export. The MRP-like family, similar to all ABC proteins, have a common four-domain core structure constituted by two membrane-spanning domains each composed of six transmembrane (TM) helices and two nucleotide-binding domains (NBD). ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213221 [Multi-domain] Cd Length: 229 Bit Score: 70.33 E-value: 7.29e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKrKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYC 1993
Cdd:cd03254 4 EFENVNFSYDEK-KPVLKDINFSIKPGETVAIVGPTGAGKTTLINLLMRFYDPQKGQILIDGIDIRDiSRKSLRSMIGVV 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQfDAIteLLTG--REHVEFFALLrgVPEKEVGKVGEWA-----IRKL--GLVKYGEKYASNYSGGNKRKLSTAMALIGG 2064
Cdd:cd03254 83 LQ-DTF--LFSGtiMENIRLGRPN--ATDEEVIEAAKEAgahdfIMKLpnGYDTVLGENGGNLSQGERQLLAIARAMLRD 157
|
170 180 190 200
....*....|....*....|....*....|....*....|
gi 568925948 2065 PPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSH 2104
Cdd:cd03254 158 PKILILDEATSNIDTETEKLIQE-ALEKLMKGRTSIIIAH 196
|
|
| PRK11000 |
PRK11000 |
maltose/maltodextrin ABC transporter ATP-binding protein MalK; |
898-1118 |
7.46e-13 |
|
maltose/maltodextrin ABC transporter ATP-binding protein MalK;
Pssm-ID: 182893 [Multi-domain] Cd Length: 369 Bit Score: 72.75 E-value: 7.46e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 898 GVSIQNLVKVYrDGMKVAVDgLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIrSEMSSIRQNLGV 977
Cdd:PRK11000 3 SVTLRNVTKAY-GDVVISKD-INLDIHEGEFVVFVGPSGCGKSTLLRMIAGLEDITSGDLFIGEKRM-NDVPPAERGVGM 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTVEEHIWFYARLKGLSEKHVKAEMEQMA--LDVGlppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:PRK11000 80 VFQSYALYPHLSVAENMSFGLKLAGAKKEEINQRVNQVAevLQLA---HLLDRKPKALSGGQRQRVAIGRTLVAEPSVFL 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1056 LDEPTAGVDPYSR---RgIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFL 1118
Cdd:PRK11000 157 LDEPLSNLDAALRvqmR-IEISRLHKRLGRTMIYVTHDQVEAMTLADKIVVLDAGRVAQVGKPLEL 221
|
|
| ABC_FeS_Assembly |
cd03217 |
ABC-type transport system involved in Fe-S cluster assembly, ATPase component; Biosynthesis of ... |
900-1114 |
7.63e-13 |
|
ABC-type transport system involved in Fe-S cluster assembly, ATPase component; Biosynthesis of iron-sulfur clusters (Fe-S) depends on multi-protein systems. The SUF system of E. coli and Erwinia chrysanthemi is important for Fe-S biogenesis under stressful conditions. The SUF system is made of six proteins: SufC is an atypical cytoplasmic ABC-ATPase, which forms a complex with SufB and SufD; SufA plays the role of a scaffold protein for assembly of iron-sulfur clusters and delivery to target proteins; SufS is a cysteine desulfurase which mobilizes the sulfur atom from cysteine and provides it to the cluster; SufE has no associated function yet.
Pssm-ID: 213184 [Multi-domain] Cd Length: 200 Bit Score: 69.48 E-value: 7.63e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLvKVYRDGMKVaVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGlFP---PTSGTAYILGKDIrSEMSS---IRQ 973
Cdd:cd03217 2 EIKDL-HVSVGGKEI-LKGVNLTIKKGEVHALMGPNGSGKSTLAKTIMG-HPkyeVTEGEILFKGEDI-TDLPPeerARL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 NLGVCPQHNVLFDMLTVEEHIWFyarlkglsekhvkaemeqmaLDVGLppsklksktsqlSGGMQRKLSVALAFVGGSKV 1053
Cdd:cd03217 78 GIFLAFQYPPEIPGVKNADFLRY--------------------VNEGF------------SGGEKKRNEILQLLLLEPDL 125
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1054 VILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEAD-ILGDRIAIISHGKLCCVGS 1114
Cdd:cd03217 126 AILDEPDSGLDIDALRLVAEVINKLReEGKSVLIITHYQRLLDyIKPDRVHVLYDGRIVKSGD 188
|
|
| PRK03695 |
PRK03695 |
vitamin B12-transporter ATPase; Provisional |
914-1114 |
8.87e-13 |
|
vitamin B12-transporter ATPase; Provisional
Pssm-ID: 235150 [Multi-domain] Cd Length: 248 Bit Score: 70.35 E-value: 8.87e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 914 VAVDG----LALNFYEGQITSFLGHNGAGKTTTMSILTGLFpPTSGTAYILGKDIR----SEMSSIRQNLgvCPQHNVLF 985
Cdd:PRK03695 6 VAVSTrlgpLSAEVRAGEILHLVGPNGAGKSTLLARMAGLL-PGSGSIQFAGQPLEawsaAELARHRAYL--SQQQTPPF 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 986 DMltveeHIWFYARL---KGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGG-MQRklsVALAFV---------GGSK 1052
Cdd:PRK03695 83 AM-----PVFQYLTLhqpDKTRTEAVASALNEVAEALGLDD-KLGRSVNQLSGGeWQR---VRLAAVvlqvwpdinPAGQ 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK03695 154 LLLLDEPMNSLDVAQQAALDRLLSELcQQGIAVVMSSHDLNHTLRHADRVWLLKQGKLLASGR 216
|
|
| PRK10982 |
PRK10982 |
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional |
911-1108 |
9.34e-13 |
|
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional
Pssm-ID: 182880 [Multi-domain] Cd Length: 491 Bit Score: 73.23 E-value: 9.34e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDML-- 988
Cdd:PRK10982 10 GVK-ALDNVNLKVRPHSIHALMGENGAGKSTLLKCLFGIYQKDSGSILFQGKEIDFKSSKEALENGISMVHQELNLVLqr 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWF--YArLKGLSEKHVKAEMEQMA----LDVGLPPsklKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:PRK10982 89 SVMDNMWLgrYP-TKGMFVDQDKMYRDTKAifdeLDIDIDP---RAKVATLSVSQMQMIEIAKAFSYNAKIVIMDEPTSS 164
|
170 180 190 200
....*....|....*....|....*....|....*....|....*..
gi 568925948 1063 VDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK10982 165 LTEKEVNHLFTIIRKLKeRGCGIVYISHKMEEIFQLCDEITILRDGQ 211
|
|
| PRK10790 |
PRK10790 |
SmdB family multidrug efflux ABC transporter permease/ATP-binding protein; |
899-1108 |
9.74e-13 |
|
SmdB family multidrug efflux ABC transporter permease/ATP-binding protein;
Pssm-ID: 182733 [Multi-domain] Cd Length: 592 Bit Score: 73.60 E-value: 9.74e-13
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDgLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:PRK10790 341 IDIDNVSFAYRDDNLVLQN-INLSVPSRGFVALVGHTGSGKSTLASLLMGYYPLTEGEIRLDGRPLSSlSHSVLRQGVAM 419
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNV-----LFDMLTVEEHIwfyarlkglSEKHVKAEME--QMALDV-GLPP---SKLKSKTSQLSGGMQRKLSVALA 1046
Cdd:PRK10790 420 VQQDPVvladtFLANVTLGRDI---------SEEQVWQALEtvQLAELArSLPDglyTPLGEQGNNLSVGQKQLLALARV 490
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1047 FVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMD---EAdilgDRIAIISHGK 1108
Cdd:PRK10790 491 LVQTPQILILDEATANIDSGTEQAIQQALAAVREHTTLVVIAHRLStivEA----DTILVLHRGQ 551
|
|
| PRK10584 |
PRK10584 |
putative ABC transporter ATP-binding protein YbbA; Provisional |
1938-2125 |
1.08e-12 |
|
putative ABC transporter ATP-binding protein YbbA; Provisional
Pssm-ID: 182569 [Multi-domain] Cd Length: 228 Bit Score: 69.81 E-value: 1.08e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSiLSNIHEVH------QNMGYCPQFDAITELLTGREHVEF 2011
Cdd:PRK10584 33 VKRGETIALIGESGSGKSTLLAILAGLDDGSSGEVSLVGQP-LHQMDEEAraklraKHVGFVFQSFMLIPTLNALENVEL 111
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2012 FALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALS 2091
Cdd:PRK10584 112 PALLRGESSRQSRNGAKALLEQLGLGKRLDHLPAQLSGGEQQRVALARAFNGRPDVLFADEPTGNLDRQTGDKIADLLFS 191
|
170 180 190
....*....|....*....|....*....|....*
gi 568925948 2092 IVKE-GRSVVLTSHSmEECEALCTRMAIMVNGRFR 2125
Cdd:PRK10584 192 LNREhGTTLILVTHD-LQLAARCDRRLRLVNGQLQ 225
|
|
| Uup |
COG0488 |
ATPase components of ABC transporters with duplicated ATPase domains [General function ... |
1911-2125 |
1.21e-12 |
|
ATPase components of ABC transporters with duplicated ATPase domains [General function prediction only];
Pssm-ID: 440254 [Multi-domain] Cd Length: 520 Bit Score: 72.79 E-value: 1.21e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilsniheVHqnM 1990
Cdd:COG0488 313 KKVLELEGLSKSY--GDKTLLDDLSLRIDRGDRIGLIGPNGAGKSTLLKLLAGELEPDSGTVKLGET--------VK--I 380
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GYCPQ-FDAITELLTGREHVEffALLRGVPEKEVGKVgewairkLGlvKYG------EKYASNYSGGNKRKLSTAMALIG 2063
Cdd:COG0488 381 GYFDQhQEELDPDKTVLDELR--DGAPGGTEQEVRGY-------LG--RFLfsgddaFKPVGVLSGGEKARLALAKLLLS 449
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2064 GPPVVFLDEPTTGMDPKARRFLwNCALsIVKEGrSVVLTSHSMEECEALCTRMAIMVNGRFR 2125
Cdd:COG0488 450 PPNVLLLDEPTNHLDIETLEAL-EEAL-DDFPG-TVLLVSHDRYFLDRVATRILEFEDGGVR 508
|
|
| phnK |
PRK11701 |
phosphonate C-P lyase system protein PhnK; Provisional |
900-1108 |
1.23e-12 |
|
phosphonate C-P lyase system protein PhnK; Provisional
Pssm-ID: 183280 [Multi-domain] Cd Length: 258 Bit Score: 70.34 E-value: 1.23e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKD---------------- 963
Cdd:PRK11701 8 SVRGLTKLY--GPRKGCRDVSFDLYPGEVLGIVGESGSGKTTLLNALSARLAPDAGEVHYRMRDgqlrdlyalseaerrr 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 964 -IRSEMSSIRQN------LGVCPQHNVlfdmltVEehiwfyaRLKGLSEKHVkAEMEQMALD----VGLPPSKLKSKTSQ 1032
Cdd:PRK11701 86 lLRTEWGFVHQHprdglrMQVSAGGNI------GE-------RLMAVGARHY-GDIRATAGDwlerVEIDAARIDDLPTT 151
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1033 LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK11701 152 FSGGMQQRLQIARNLVTHPRLVFMDEPTGGLDVSVQARLLDLLrgLVRELGLAVVIVTHDLAVARLLAHRLLVMKQGR 229
|
|
| cbiO |
PRK13648 |
cobalt transporter ATP-binding subunit; Provisional |
1909-2121 |
1.27e-12 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184207 [Multi-domain] Cd Length: 269 Bit Score: 70.55 E-value: 1.27e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1909 GQNDILEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI----LSNIH 1984
Cdd:PRK13648 3 DKNSIIVFKNVSFQYQSDASFTLKDVSFNIPKGQWTSIVGHNGSGKSTIAKLMIGIEKVKSGEIFYNNQAItddnFEKLR 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 E----VHQN-----MGYCPQFDaitelltgrehVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKL 2055
Cdd:PRK13648 83 KhigiVFQNpdnqfVGSIVKYD-----------VAFGLENHAVPYDEMHRRVSEALKQVDMLERADYEPNALSGGQKQRV 151
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2056 STAMALIGGPPVVFLDEPTTGMDPKARRFLWNCaLSIVKEGRSVVLTSHSMEECEALCTRMAIMVN 2121
Cdd:PRK13648 152 AIAGVLALNPSVIILDEATSMLDPDARQNLLDL-VRKVKSEHNITIISITHDLSEAMEADHVIVMN 216
|
|
| potA |
PRK09452 |
spermidine/putrescine ABC transporter ATP-binding protein PotA; |
1913-2137 |
1.28e-12 |
|
spermidine/putrescine ABC transporter ATP-binding protein PotA;
Pssm-ID: 236523 [Multi-domain] Cd Length: 375 Bit Score: 71.90 E-value: 1.28e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS------NIHEV 1986
Cdd:PRK09452 14 LVELRGISKSF--DGKEVISNLDLTINNGEFLTLLGPSGCGKTTVLRLIAGFETPDSGRIMLDGQDITHvpaenrHVNTV 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPP 2066
Cdd:PRK09452 92 FQSYALFPH-------MTVFENVAFGLRMQKTPAAEITPRVMEALRMVQLEEFAQRKPHQLSGGQQQRVAIARAVVNKPK 164
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2067 VVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL----KNRF 2137
Cdd:PRK09452 165 VLLLDESLSALDYKLRKQMQNELKALQRKlGITFVFVTHDQEEALTMSDRIVVMRDGRIEQDGTPREIyeepKNLF 240
|
|
| AztA |
NF040873 |
zinc ABC transporter ATP-binding protein AztA; |
1929-2108 |
1.32e-12 |
|
zinc ABC transporter ATP-binding protein AztA;
Pssm-ID: 468810 [Multi-domain] Cd Length: 191 Bit Score: 68.41 E-value: 1.32e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknsilSNIHEVHQNMGYCPQFDAITELL--TGR 2006
Cdd:NF040873 6 PVLHGVDLTIPAGSLTAVVGPNGSGKSTLLKVLAGVLRPTSG----------TVRRAGGARVAYVPQRSEVPDSLplTVR 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2007 EHVE--FFA---LLRGvPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKA 2081
Cdd:NF040873 76 DLVAmgRWArrgLWRR-LTRDDRAAVDDALERVGLADLAGRQLGELSGGQRQRALLAQGLAQEADLLLLDEPTTGLDAES 154
|
170 180
....*....|....*....|....*..
gi 568925948 2082 RRFLWNCALSIVKEGRSVVLTSHSMEE 2108
Cdd:NF040873 155 RERIIALLAEEHARGATVVVVTHDLEL 181
|
|
| ArpD |
COG4618 |
ABC-type protease/lipase transport system, ATPase and permease components [Intracellular ... |
889-1109 |
1.42e-12 |
|
ABC-type protease/lipase transport system, ATPase and permease components [Intracellular trafficking, secretion, and vesicular transport];
Pssm-ID: 443660 [Multi-domain] Cd Length: 563 Bit Score: 72.86 E-value: 1.42e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHL---RLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR 965
Cdd:COG4618 318 EPERMPLprpKGRLSVENLTVVPPGSKRPILRGVSFSLEPGEVLGVIGPSGSGKSTLARLLVGVWPPTAGSVRLDGADLS 397
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 966 S-EMSSIRQNLGVCPQHNVLFDMlTVEEHIwfyARLKGL-SEKHVKAEmeQMA----LDVGLP---PSKLKSKTSQLSGG 1036
Cdd:COG4618 398 QwDREELGRHIGYLPQDVELFDG-TIAENI---ARFGDAdPEKVVAAA--KLAgvheMILRLPdgyDTRIGEGGARLSGG 471
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1037 -MQRklsVALA--FVGGSKVVILDEPTAGVDPYSRRGIWELL--LKyRQGRTIILSTHHMdeaDILG--DRIAIISHGKL 1109
Cdd:COG4618 472 qRQR---IGLAraLYGDPRLVVLDEPNSNLDDEGEAALAAAIraLK-ARGATVVVITHRP---SLLAavDKLLVLRDGRV 544
|
|
| PRK09700 |
PRK09700 |
D-allose ABC transporter ATP-binding protein AlsA; |
1881-2123 |
1.44e-12 |
|
D-allose ABC transporter ATP-binding protein AlsA;
Pssm-ID: 182036 [Multi-domain] Cd Length: 510 Bit Score: 72.51 E-value: 1.44e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1881 RPVKAKLPPLNDEDEDVRRERqrildgggqndILEIKELTkiyRRKRKPAVDrICIGIPPGECFGLLGVNGAGKSTTFKM 1960
Cdd:PRK09700 244 RELQNRFNAMKENVSNLAHET-----------VFEVRNVT---SRDRKKVRD-ISFSVCRGEILGFAGLVGSGRTELMNC 308
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1961 LTGDTPVTRGDAFLN--KNSILSNIHEVHQNMGYcpqfdaITElltGREHVEFF---------ALLRGVpeKEVGKVGEW 2029
Cdd:PRK09700 309 LFGVDKRAGGEIRLNgkDISPRSPLDAVKKGMAY------ITE---SRRDNGFFpnfsiaqnmAISRSL--KDGGYKGAM 377
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2030 AI-------------RKLGLVKYG--EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVK 2094
Cdd:PRK09700 378 GLfhevdeqrtaenqRELLALKCHsvNQNITELSGGNQQKVLISKWLCCCPEVIIFDEPTRGIDVGAKAEIYKVMRQLAD 457
|
250 260
....*....|....*....|....*....
gi 568925948 2095 EGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK09700 458 DGKVILMVSSELPEIITVCDRIAVFCEGR 486
|
|
| CydD |
TIGR02857 |
thiol reductant ABC exporter, CydD subunit; The gene pair cydCD encodes an ABC-family ... |
1914-2107 |
1.57e-12 |
|
thiol reductant ABC exporter, CydD subunit; The gene pair cydCD encodes an ABC-family transporter in which each gene contains an N-terminal membrane-spanning domain (pfam00664) and a C-terminal ATP-binding domain (pfam00005). In E. coli these genes were discovered as mutants which caused the terminal heme-copper oxidase complex cytochrome bd to fail to assemble. Recent work has shown that the transporter is involved in export of redox-active thiol compounds such as cysteine and glutathione. The linkage to assembly of the cytochrome bd complex is further supported by the conserved operon structure found outside the gammaproteobacteria (cydABCD) containing both the transporter and oxidase genes components. The genes used as the seed members for this model are all either found in the gammproteobacterial context or the CydABCD context. All members of this family scoring above trusted at the time of its creation were from genomes which encode a cytochrome bd complex. Unfortunately, the gene symbol nomenclature adopted based on this operon in B. subtilis assigns cydC to the third gene in the operon where this gene is actually homologous to the E. coli cydD gene. We have chosen to name all homologs in this family in accordance with the precedence of publication of the E. coli name, CydD
Pssm-ID: 274323 [Multi-domain] Cd Length: 529 Bit Score: 72.70 E-value: 1.57e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGY 1992
Cdd:TIGR02857 322 LEFSGVSVAYPGRR-PALRPVSFTVPPGERVALVGPSGAGKSTLLNLLLGFVDPTEGSIAVNGVPLADaDADSWRDQIAW 400
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQFDAITE------LLTGR------------EHVEFFALLRGVPEKEVGKVGEwaiRKLGLvkygekyasnySGGNKRK 2054
Cdd:TIGR02857 401 VPQHPFLFAgtiaenIRLARpdasdaeirealERAGLDEFVAALPQGLDTPIGE---GGAGL-----------SGGQAQR 466
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2055 LSTAMALIGGPPVVFLDEPTTGMDPKARRFLwNCALSIVKEGRSVVLTSHSME 2107
Cdd:TIGR02857 467 LALARAFLRDAPLLLLDEPTAHLDAETEAEV-LEALRALAQGRTVLLVTHRLA 518
|
|
| PRK13549 |
PRK13549 |
xylose transporter ATP-binding subunit; Provisional |
902-1108 |
1.61e-12 |
|
xylose transporter ATP-binding subunit; Provisional
Pssm-ID: 184134 [Multi-domain] Cd Length: 506 Bit Score: 72.65 E-value: 1.61e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 902 QNLVKVYrDGMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP--PTSGTAYILGKDIR------SE---MSS 970
Cdd:PRK13549 9 KNITKTF-GGVK-ALDNVSLKVRAGEIVSLCGENGAGKSTLMKVLSGVYPhgTYEGEIIFEGEELQasnirdTEragIAI 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQNLGVCPQhnvlfdmLTVEEHI-----WFYARLKGLSEKHVKAE--MEQMALDVglPPSklkSKTSQLSGGMQRKLSV 1043
Cdd:PRK13549 87 IHQELALVKE-------LSVLENIflgneITPGGIMDYDAMYLRAQklLAQLKLDI--NPA---TPVGNLGLGQQQLVEI 154
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKyRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK13549 155 AKALNKQARLLILDEPTASLTESETAVLLDIIrdLK-AHGIACIYISHKLNEVKAISDTICVIRDGR 220
|
|
| PRK13549 |
PRK13549 |
xylose transporter ATP-binding subunit; Provisional |
916-1109 |
1.80e-12 |
|
xylose transporter ATP-binding subunit; Provisional
Pssm-ID: 184134 [Multi-domain] Cd Length: 506 Bit Score: 72.27 E-value: 1.80e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP-PTSGTAYILGK--DIRSEMSSIRQNLGVCP----QHNVLFDMl 988
Cdd:PRK13549 278 VDDVSFSLRRGEILGIAGLVGAGRTELVQCLFGAYPgRWEGEIFIDGKpvKIRNPQQAIAQGIAMVPedrkRDGIVPVM- 356
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWF--YARLKGLSekHVKAEMEQMALDVGLppSKLKSKTS-------QLSGGMQRKLSVALAFVGGSKVVILDEP 1059
Cdd:PRK13549 357 GVGKNITLaaLDRFTGGS--RIDDAAELKTILESI--QRLKVKTAspelaiaRLSGGNQQKAVLAKCLLLNPKILILDEP 432
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1060 TAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEadILG--DRIAIISHGKL 1109
Cdd:PRK13549 433 TRGIDVGAKYEIYKLINQLvQQGVAIIVISSELPE--VLGlsDRVLVMHEGKL 483
|
|
| ThiQ |
COG3840 |
ABC-type thiamine transport system, ATPase component ThiQ [Coenzyme transport and metabolism]; |
1914-2123 |
1.86e-12 |
|
ABC-type thiamine transport system, ATPase component ThiQ [Coenzyme transport and metabolism];
Pssm-ID: 443051 [Multi-domain] Cd Length: 232 Bit Score: 69.01 E-value: 1.86e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKpavdRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHE------- 1985
Cdd:COG3840 2 LRLDDLTYRYGDFPL----RFDLTIAAGERVAILGPSGAGKSTLLNLIAGFLPPDSGRILWNGQDLTAlPPAErpvsmlf 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 ----------VHQNMGycpqfdaitelltgrehvefFAL---LR-GVPEKEvgKVgEWAIRKLGLVKYGEKYASNYSGGN 2051
Cdd:COG3840 78 qennlfphltVAQNIG--------------------LGLrpgLKlTAEQRA--QV-EQALERVGLAGLLDRLPGQLSGGQ 134
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2052 KRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLwncaLSIVKE-----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG3840 135 RQRVALARCLVRKRPILLLDEPFSALDPALRQEM----LDLVDElcrerGLTVLMVTHDPEDAARIADRVLLVADGR 207
|
|
| COG4559 |
COG4559 |
ABC-type hemin transport system, ATPase component [Inorganic ion transport and metabolism]; |
1914-2131 |
2.37e-12 |
|
ABC-type hemin transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443620 [Multi-domain] Cd Length: 258 Bit Score: 69.37 E-value: 2.37e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-------LSNIHEV 1986
Cdd:COG4559 2 LEAENLS--VRLGGRTLLDDVSLTLRPGELTAIIGPNGAGKSTLLKLLTGELTPSSGEVRLNGRPLaawspweLARRRAV 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HqnmgycPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALI---- 2062
Cdd:COG4559 80 L------PQHSSLAFPFTVEEVVALGRAPHGSSAAQDRQIVREALALVGLAHLAGRSYQTLSGGEQQRVQLARVLAqlwe 153
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2063 ---GGPPVVFLDEPTTGMDPK--------ARRFlwncalsiVKEGRSVV-------LTSHsmeeceaLCTRMAIMVNGRF 2124
Cdd:COG4559 154 pvdGGPRWLFLDEPTSALDLAhqhavlrlARQL--------ARRGGGVVavlhdlnLAAQ-------YADRILLLHQGRL 218
|
....*..
gi 568925948 2125 RCLGSVQ 2131
Cdd:COG4559 219 VAQGTPE 225
|
|
| cbiO |
PRK13644 |
energy-coupling factor transporter ATPase; |
1929-2193 |
2.44e-12 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 106587 [Multi-domain] Cd Length: 274 Bit Score: 69.63 E-value: 2.44e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN--KNSILSNIHEVHQNMGYCPQfDAITELL--T 2004
Cdd:PRK13644 16 PALENINLVIKKGEYIGIIGKNGSGKSTLALHLNGLLRPQKGKVLVSgiDTGDFSKLQGIRKLVGIVFQ-NPETQFVgrT 94
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2005 GREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRF 2084
Cdd:PRK13644 95 VEEDLAFGPENLCLPPIEIRKRVDRALAEIGLEKYRHRSPKTLSGGQGQCVALAGILTMEPECLIFDEVTSMLDPDSGIA 174
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2085 LWNCALSIVKEGRSVVLTSHSMEECEAlCTRMAIMVNGRFRCLGSVQHLKnrfgdgytivvriagSNPDLKpvqeFFGLA 2164
Cdd:PRK13644 175 VLERIKKLHEKGKTIVYITHNLEELHD-ADRIIVMDRGKIVLEGEPENVL---------------SDVSLQ----TLGLT 234
|
250 260 270
....*....|....*....|....*....|....*..
gi 568925948 2165 FPGSV-LKE---KHRNMLQYQLPSSLSSLA----RIF 2193
Cdd:PRK13644 235 PPSLIeLAEnlkMHGVVIPWENTSSPSSFAeeicRLF 271
|
|
| livG |
PRK11300 |
leucine/isoleucine/valine transporter ATP-binding subunit; Provisional |
1930-2123 |
2.79e-12 |
|
leucine/isoleucine/valine transporter ATP-binding subunit; Provisional
Pssm-ID: 183080 [Multi-domain] Cd Length: 255 Bit Score: 69.25 E-value: 2.79e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI--LSNiHEV--------HQNMGYCPQFDAI 1999
Cdd:PRK11300 20 AVNNVNLEVREQEIVSLIGPNGAGKTTVFNCLTGFYKPTGGTILLRGQHIegLPG-HQIarmgvvrtFQHVRLFREMTVI 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2000 TELLTGR-EHVE--FFALLRGVP-----EKE-VGKVGEWaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:PRK11300 99 ENLLVAQhQQLKtgLFSGLLKTPafrraESEaLDRAATW-LERVGLLEHANRQAGNLAYGQQRRLEIARCMVTQPEILML 177
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK11300 178 DEPAAGLNPKETKELDELIAELRNEhNVTVLLIEHDMKLVMGISDRIYVVNQGT 231
|
|
| 3a01205 |
TIGR00956 |
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other] |
1889-2104 |
3.14e-12 |
|
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273362 [Multi-domain] Cd Length: 1394 Bit Score: 72.45 E-value: 3.14e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1889 PLNDEDEDVRRERQRILDGGGqnDILEIKELT---KIYRRKRK--PAVDRICIgipPGECFGLLGVNGAGKSTTFKMLTG 1963
Cdd:TIGR00956 737 DLTDESDDVNDEKDMEKESGE--DIFHWRNLTyevKIKKEKRVilNNVDGWVK---PGTLTALMGASGAGKTTLLNVLAE 811
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1964 DTP---VTRGDAFLNKNSILSNIHevhQNMGYCPQFDAITELLTGREHVEFFALLR---GVPEKEVGKVGEWAIRKLGLV 2037
Cdd:TIGR00956 812 RVTtgvITGGDRLVNGRPLDSSFQ---RSIGYVQQQDLHLPTSTVRESLRFSAYLRqpkSVSKSEKMEYVEEVIKLLEME 888
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2038 KYGEKYASNYSGG----NKRKLSTAMALIGGPP-VVFLDEPTTGMDPKArrflwncALSIVK-------EGRSVVLTSH 2104
Cdd:TIGR00956 889 SYADAVVGVPGEGlnveQRKRLTIGVELVAKPKlLLFLDEPTSGLDSQT-------AWSICKlmrkladHGQAILCTIH 960
|
|
| PRK14267 |
PRK14267 |
phosphate ABC transporter ATP-binding protein; Provisional |
1938-2105 |
3.43e-12 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 184596 [Multi-domain] Cd Length: 253 Bit Score: 68.72 E-value: 3.43e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKST---TFKMLTGDTPVTR--GDAFLNKNSILS---NIHEVHQNMGYCPQFDAITELLTGREHV 2009
Cdd:PRK14267 27 IPQNGVFALMGPSGCGKSTllrTFNRLLELNEEARveGEVRLFGRNIYSpdvDPIEVRREVGMVFQYPNPFPHLTIYDNV 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2010 EFFALLRGV--PEKEVGKVGEWAIRKLGL---VKYGEK-YASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARR 2083
Cdd:PRK14267 107 AIGVKLNGLvkSKKELDERVEWALKKAALwdeVKDRLNdYPSNLSGGQRQRLVIARALAMKPKILLMDEPTANIDPVGTA 186
|
170 180
....*....|....*....|..
gi 568925948 2084 FLWNCALSIvKEGRSVVLTSHS 2105
Cdd:PRK14267 187 KIEELLFEL-KKEYTIVLVTHS 207
|
|
| ArtP |
COG4161 |
ABC-type arginine transport system, ATPase component [Amino acid transport and metabolism]; |
897-1109 |
3.99e-12 |
|
ABC-type arginine transport system, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 443326 [Multi-domain] Cd Length: 242 Bit Score: 68.50 E-value: 3.99e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-------RSEMS 969
Cdd:COG4161 1 MSIQLKNINCFY--GSHQALFDINLECPSGETLVLLGPSGAGKSSLLRVLNLLETPDSGQLNIAGHQFdfsqkpsEKAIR 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 970 SIRQNLGVCPQHNVLFDMLTVEEH-IWFYARLKGLSEKHVKAEMEQMALDVGLPPsKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:COG4161 79 LLRQKVGMVFQQYNLWPHLTVMENlIEAPCKVLGLSKEQAREKAMKLLARLRLTD-KADRFPLHLSGGQQQRVAIARALM 157
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:COG4161 158 MEPQVLLFDEPTAALDPEITAQVVEIIRELSQtGITQVIVTHEVEFARKVASQVVYMEKGRI 219
|
|
| PhnK |
COG1101 |
ABC-type uncharacterized transport system, ATPase component [General function prediction only]; ... |
1913-2123 |
5.41e-12 |
|
ABC-type uncharacterized transport system, ATPase component [General function prediction only];
Pssm-ID: 440718 [Multi-domain] Cd Length: 264 Bit Score: 68.57 E-value: 5.41e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKR---KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL--------S 1981
Cdd:COG1101 1 MLELKNLSKTFNPGTvneKRALDGLNLTIEEGDFVTVIGSNGAGKSTLLNAIAGSLPPDSGSILIDGKDVTklpeykraK 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1982 NIHEVHQN--MGYCPQfdaitelLTGREHV-------EFFALLRGVPEKEVGKVGEW-AIRKLGLvkygEKY----ASNY 2047
Cdd:COG1101 81 YIGRVFQDpmMGTAPS-------MTIEENLalayrrgKRRGLRRGLTKKRRELFRELlATLGLGL----ENRldtkVGLL 149
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2048 SGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRsvvLTS----HSMEECEALCTRMAIMVNGR 2123
Cdd:COG1101 150 SGGQRQALSLLMATLTKPKLLLLDEHTAALDPKTAALVLELTEKIVEENN---LTTlmvtHNMEQALDYGNRLIMMHEGR 226
|
|
| NupO |
COG3845 |
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and ... |
1913-2123 |
6.38e-12 |
|
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and metabolism];
Pssm-ID: 443055 [Multi-domain] Cd Length: 504 Bit Score: 70.44 E-value: 6.38e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrkrkPAV---DRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN---------KNSIL 1980
Cdd:COG3845 5 ALELRGITKRF-----GGVvanDDVSLTVRPGEIHALLGENGAGKSTLMKILYGLYQPDSGEILIDgkpvrirspRDAIA 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1981 SNIHEVHQN-MgycpQFDAIT--E-LLTGREHVEFFALLRGVPEKEVGKVGEwairklglvKYG-----EKYASNYSGGN 2051
Cdd:COG3845 80 LGIGMVHQHfM----LVPNLTvaEnIVLGLEPTKGGRLDRKAARARIRELSE---------RYGldvdpDAKVEDLSVGE 146
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2052 KRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG3845 147 QQRVEILKALYRGARILILDEPTAVLTPQEADELFEILRRLAAEGKSIIFITHKLREVMAIADRVTVLRRGK 218
|
|
| PRK15056 |
PRK15056 |
manganese/iron ABC transporter ATP-binding protein; |
1910-2122 |
7.34e-12 |
|
manganese/iron ABC transporter ATP-binding protein;
Pssm-ID: 185016 [Multi-domain] Cd Length: 272 Bit Score: 68.37 E-value: 7.34e-12
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAflnknSIL--SNIHEVH 1987
Cdd:PRK15056 3 QQAGIVVNDVTVTWRNGHT-ALRDASFTVPGGSIAALVGVNGSGKSTLFKALMGFVRLASGKI-----SILgqPTRQALQ 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNM-GYCPQ-------FDAITE--LLTGRE-HVeffALLRgVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLS 2056
Cdd:PRK15056 77 KNLvAYVPQseevdwsFPVLVEdvVMMGRYgHM---GWLR-RAKKRDRQIVTAALARVDMVEFRHRQIGELSGGQKKRVF 152
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2057 TAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTrMAIMVNG 2122
Cdd:PRK15056 153 LARAIAQQGQVILLDEPFTGVDVKTEARIISLLRELRDEGKTMLVSTHNLGSVTEFCD-YTVMVKG 217
|
|
| PRK10253 |
PRK10253 |
iron-enterobactin ABC transporter ATP-binding protein; |
888-1114 |
1.17e-11 |
|
iron-enterobactin ABC transporter ATP-binding protein;
Pssm-ID: 182336 [Multi-domain] Cd Length: 265 Bit Score: 67.32 E-value: 1.17e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 888 MEEEPTHLRLgvsiQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE 967
Cdd:PRK10253 1 MTESVARLRG----EQLTLGY--GKYTVAENLTVEIPDGHFTAIIGPNGCGKSTLLRTLSRLMTPAHGHVWLDGEHIQHY 74
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MS-SIRQNLGVCPQHNVLFDMLTVEE--------HIWFYARLKGLSEKHVKAEMEQMALdvglppSKLKSKT-SQLSGGM 1037
Cdd:PRK10253 75 ASkEVARRIGLLAQNATTPGDITVQElvargrypHQPLFTRWRKEDEEAVTKAMQATGI------THLADQSvDTLSGGQ 148
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1038 QRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK10253 149 RQRAWIAMVLAQETAIMLLDEPTTWLDISHQIDLLELLseLNREKGYTLAAVLHDLNQACRYASHLIALREGKIVAQGA 227
|
|
| PRK13651 |
PRK13651 |
cobalt transporter ATP-binding subunit; Provisional |
1914-2107 |
1.22e-11 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 184210 [Multi-domain] Cd Length: 305 Bit Score: 68.19 E-value: 1.22e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRK---PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG------------------DTPVTRGDA 1972
Cdd:PRK13651 3 IKVKNIVKIFNKKLPtelKALDNVSVEINQGEFIAIIGQTGSGKTTFIEHLNAlllpdtgtiewifkdeknKKKTKEKEK 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1973 FLNKNSI-------LSNIHEVHQNMGYCPQFdAITELL--TGREHVEFFALLRGVPEKEVGKVGEWAIRKLGL-VKYGEK 2042
Cdd:PRK13651 83 VLEKLVIqktrfkkIKKIKEIRRRVGVVFQF-AEYQLFeqTIEKDIIFGPVSMGVSKEEAKKRAAKYIELVGLdESYLQR 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2043 YASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSME 2107
Cdd:PRK13651 162 SPFELSGGQKRRVALAGILAMEPDFLVFDEPTAGLDPQGVKEILEIFDNLNKQGKTIILVTHDLD 226
|
|
| PRK15056 |
PRK15056 |
manganese/iron ABC transporter ATP-binding protein; |
898-1091 |
1.43e-11 |
|
manganese/iron ABC transporter ATP-binding protein;
Pssm-ID: 185016 [Multi-domain] Cd Length: 272 Bit Score: 67.22 E-value: 1.43e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 898 GVSIQNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSemsSIRQNL-G 976
Cdd:PRK15056 6 GIVVNDVTVTWRNG-HTALRDASFTVPGGSIAALVGVNGSGKSTLFKALMGFVRLASGKISILGQPTRQ---ALQKNLvA 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHN-------VLFD---MLTVEEHIWFYARLKGLSEKHVKAEMEQmaldVGLPPSKLKsKTSQLSGGMQRKLSVALA 1046
Cdd:PRK15056 82 YVPQSEevdwsfpVLVEdvvMMGRYGHMGWLRRAKKRDRQIVTAALAR----VDMVEFRHR-QIGELSGGQKKRVFLARA 156
|
170 180 190 200
....*....|....*....|....*....|....*....|....*.
gi 568925948 1047 FVGGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHM 1091
Cdd:PRK15056 157 IAQQGQVILLDEPFTGVDVKTEARIISLLRELRdEGKTMLVSTHNL 202
|
|
| MK0520 |
COG2401 |
ABC-type ATPase fused to a predicted acetyltransferase domain [General function prediction ... |
896-1107 |
1.73e-11 |
|
ABC-type ATPase fused to a predicted acetyltransferase domain [General function prediction only];
Pssm-ID: 441957 [Multi-domain] Cd Length: 222 Bit Score: 66.14 E-value: 1.73e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 896 RLGVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTG--LFPPTSGTAYILGKDIRSEMSSIrq 973
Cdd:COG2401 26 RVAIVLEAFGVELRVVERYVLRDLNLEIEPGEIVLIVGASGSGKSTLLRLLAGalKGTPVAGCVDVPDNQFGREASLI-- 103
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 nlgvcpqhnvlfdmltveEHIWfyarlkglSEKHVKAEMEQMAlDVGL--PPSkLKSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:COG2401 104 ------------------DAIG--------RKGDFKDAVELLN-AVGLsdAVL-WLRRFKELSTGQKFRFRLALLLAERP 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1052 KVVILDEPTAGVDP-YSRRG--IWELLLKyRQGRTIILSTHHMD-EADILGDRIAIISHG 1107
Cdd:COG2401 156 KLLVIDEFCSHLDRqTAKRVarNLQKLAR-RAGITLVVATHHYDvIDDLQPDLLIFVGYG 214
|
|
| ABCF_EF-3 |
cd03221 |
ATP-binding cassette domain of elongation factor 3, subfamily F; Elongation factor 3 (EF-3) is ... |
899-1108 |
2.15e-11 |
|
ATP-binding cassette domain of elongation factor 3, subfamily F; Elongation factor 3 (EF-3) is a cytosolic protein required by fungal ribosomes for in vitro protein synthesis and for in vivo growth. EF-3 stimulates the binding of the EF-1: GTP: aa-tRNA ternary complex to the ribosomal A site by facilitated release of the deacylated tRNA from the E site. The reaction requires ATP hydrolysis. EF-3 contains two ATP nucleotide binding sequence (NBS) motifs. NBSI is sufficient for the intrinsic ATPase activity. NBSII is essential for the ribosome-stimulated functions.
Pssm-ID: 213188 [Multi-domain] Cd Length: 144 Bit Score: 63.62 E-value: 2.15e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayilgkdirsemssirqnlgvc 978
Cdd:cd03221 1 IELENLSKTYGG--KLLLKDISLTINPGDRIGLVGRNGAGKSTLLKLIAGELEPDEGI---------------------- 56
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 pqhnvlfdmltveehiwfYARLKGLSEKHVkaemeqmaldvglppsklksktSQLSGGMQRKLSVALAFVGGSKVVILDE 1058
Cdd:cd03221 57 ------------------VTWGSTVKIGYF----------------------EQLSGGEKMRLALAKLLLENPNLLLLDE 96
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1059 PTAGVDPYSRRGIWELLLKYRqgRTIILSTHhmDEA--DILGDRIAIISHGK 1108
Cdd:cd03221 97 PTNHLDLESIEALEEALKEYP--GTVILVSH--DRYflDQVATKIIELEDGK 144
|
|
| PRK15079 |
PRK15079 |
oligopeptide ABC transporter ATP-binding protein OppF; Provisional |
915-1064 |
2.61e-11 |
|
oligopeptide ABC transporter ATP-binding protein OppF; Provisional
Pssm-ID: 185037 [Multi-domain] Cd Length: 331 Bit Score: 67.42 E-value: 2.61e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR----SEMSSIRQNLGVCPQHNV--LFDML 988
Cdd:PRK15079 36 AVDGVTLRLYEGETLGVVGESGCGKSTFARAIIGLVKATDGEVAWLGKDLLgmkdDEWRAVRSDIQMIFQDPLasLNPRM 115
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHI-----WFYARlkgLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGV 1063
Cdd:PRK15079 116 TIGEIIaeplrTYHPK---LSRQEVKDRVKAMMLKVGLLPNLINRYPHEFSGGQCQRIGIARALILEPKLIICDEPVSAL 192
|
.
gi 568925948 1064 D 1064
Cdd:PRK15079 193 D 193
|
|
| ABCG_PDR_domain2 |
cd03232 |
Second domain of the pleiotropic drug resistance-like (PDR) subfamily G of ATP-binding ... |
926-1089 |
2.64e-11 |
|
Second domain of the pleiotropic drug resistance-like (PDR) subfamily G of ATP-binding cassette transporters; The pleiotropic drug resistance (PDR) is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. This PDR subfamily represents domain I of its (ABC-IM)2 organization. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds including sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213199 [Multi-domain] Cd Length: 192 Bit Score: 64.96 E-value: 2.64e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSIL-----TGLfppTSGTAYILGKDIRSemsSIRQNLGVCPQHNVLFDMLTVEEHIWFYARL 1000
Cdd:cd03232 33 GTLTALMGESGAGKTTLLDVLagrktAGV---ITGEILINGRPLDK---NFQRSTGYVEQQDVHSPNLTVREALRFSALL 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 KGLSekhvkaeMEQmaldvglppsklksktsqlsggmQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKY-R 1079
Cdd:cd03232 107 RGLS-------VEQ-----------------------RKRLTIGVELAAKPSILFLDEPTSGLDSQAAYNIVRFLKKLaD 156
|
170
....*....|
gi 568925948 1080 QGRTIILSTH 1089
Cdd:cd03232 157 SGQAILCTIH 166
|
|
| araG |
PRK11288 |
L-arabinose ABC transporter ATP-binding protein AraG; |
926-1109 |
2.81e-11 |
|
L-arabinose ABC transporter ATP-binding protein AraG;
Pssm-ID: 183077 [Multi-domain] Cd Length: 501 Bit Score: 68.40 E-value: 2.81e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK--DIRSEMSSIRQNLGVCPQ---HNVLFDMLTVEEHIWFYARL 1000
Cdd:PRK11288 279 GEIVGLFGLVGAGRSELMKLLYGATRRTAGQVYLDGKpiDIRSPRDAIRAGIMLCPEdrkAEGIIPVHSVADNINISARR 358
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 KGLSEKHV---KAEMEQMALDVGlppsKLKSKTS-------QLSGGMQRK--LSVALAfvGGSKVVILDEPTAGVDPYSR 1068
Cdd:PRK11288 359 HHLRAGCLinnRWEAENADRFIR----SLNIKTPsreqlimNLSGGNQQKaiLGRWLS--EDMKVILLDEPTRGIDVGAK 432
|
170 180 190 200
....*....|....*....|....*....|....*....|....
gi 568925948 1069 RGIWELLLKY-RQGRTIILSTHhmDEADILG--DRIAIISHGKL 1109
Cdd:PRK11288 433 HEIYNVIYELaAQGVAVLFVSS--DLPEVLGvaDRIVVMREGRI 474
|
|
| cbiO |
PRK13641 |
energy-coupling factor transporter ATPase; |
1928-2123 |
3.06e-11 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 237456 [Multi-domain] Cd Length: 287 Bit Score: 66.78 E-value: 3.06e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRG-----DAFLNKNSILSNIHEVHQNMGYCPQFDAiTEL 2002
Cdd:PRK13641 20 KKGLDNISFELEEGSFVALVGHTGSGKSTLMQHFNALLKPSSGtitiaGYHITPETGNKNLKKLRKKVSLVFQFPE-AQL 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2003 L--TGREHVEFFALLRGVPEKEV-GKVGEWaIRKLGLVK-YGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK13641 99 FenTVLKDVEFGPKNFGFSEDEAkEKALKW-LKKVGLSEdLISKSPFELSGGQMRRVAIAGVMAYEPEILCLDEPAAGLD 177
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 2079 PKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13641 178 PEGRKEMMQLFKDYQKAGHTVILVTHNMDDVAEYADDVLVLEHGK 222
|
|
| artP |
PRK11124 |
arginine transporter ATP-binding subunit; Provisional |
899-1065 |
3.21e-11 |
|
arginine transporter ATP-binding subunit; Provisional
Pssm-ID: 182980 [Multi-domain] Cd Length: 242 Bit Score: 65.81 E-value: 3.21e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILG-----------KDIRSe 967
Cdd:PRK11124 3 IQLNGINCFY--GAHQALFDITLDCPQGETLVLLGPSGAGKSSLLRVLNLLEMPRSGTLNIAGnhfdfsktpsdKAIRE- 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 mssIRQNLGVCPQHNVLFDMLTVEEH-IWFYARLKGLSEKHVKAEMEQMaldvgLPPSKLKSKTS----QLSGGMQRKLS 1042
Cdd:PRK11124 80 ---LRRNVGMVFQQYNLWPHLTVQQNlIEAPCRVLGLSKDQALARAEKL-----LERLRLKPYADrfplHLSGGQQQRVA 151
|
170 180
....*....|....*....|...
gi 568925948 1043 VALAFVGGSKVVILDEPTAGVDP 1065
Cdd:PRK11124 152 IARALMMEPQVLLFDEPTAALDP 174
|
|
| ABC2_membrane_3 |
pfam12698 |
ABC-2 family transporter protein; This family is related to the ABC-2 membrane transporter ... |
640-841 |
3.42e-11 |
|
ABC-2 family transporter protein; This family is related to the ABC-2 membrane transporter family pfam01061.
Pssm-ID: 463674 [Multi-domain] Cd Length: 345 Bit Score: 67.03 E-value: 3.42e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 640 MPLFMTLAWIYSVAVIIKSIVYEKEARLKETMRIMGLDNGILWFSWFVSSLIPLLVSAGLLVVILkLGNLLPYSDPSVVF 719
Cdd:pfam12698 164 VGLILMIIILIGAAIIAVSIVEEKESRIKERLLVSGVSPLQYWLGKILGDFLVGLLQLLIILLLL-FGIGIPFGNLGLLL 242
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 720 VFLSVFAMVTILQCFLISTLFSRANLAAACGGIIYFTLYLPYVLcVAWQDYVGFSIKIFASLLSPVAFGFGceyfalFEE 799
Cdd:pfam12698 243 LLFLLYGLAYIALGYLLGSLFKNSEDAQSIIGIVILLLSGFFGG-LFPLEDPPSFLQWIFSIIPFFSPIDG------LLR 315
|
170 180 190 200
....*....|....*....|....*....|....*....|..
gi 568925948 800 QGIGVQWDNLFESpveedgfnlttaVSMMLFDTFLYGVMTWY 841
Cdd:pfam12698 316 LIYGDSLWEIAPS------------LIILLLFAVVLLLLALL 345
|
|
| ABC2_perm_RbbA |
NF033858 |
ribosome-associated ATPase/putative transporter RbbA; |
1930-2110 |
3.90e-11 |
|
ribosome-associated ATPase/putative transporter RbbA;
Pssm-ID: 468210 [Multi-domain] Cd Length: 907 Bit Score: 68.61 E-value: 3.90e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTF-------KMLTGDTPVTRGDaflnknsILSNIH--EVHQNMGYCPQfdait 2000
Cdd:NF033858 16 ALDDVSLDIPAGCMVGLIGPDGVGKSSLLsliagarKIQQGRVEVLGGD-------MADARHrrAVCPRIAYMPQ----- 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 EL-------LTGREHVEFFALLRGVPEKEvgkvGEWAIRKL----GLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:NF033858 84 GLgknlyptLSVFENLDFFGRLFGQDAAE----RRRRIDELlratGLAPFADRPAGKLSGGMKQKLGLCCALIHDPDLLI 159
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 2070 LDEPTTGMDPKARRFLWNCALSIVKE--GRSVVLTSHSMEECE 2110
Cdd:NF033858 160 LDEPTTGVDPLSRRQFWELIDRIRAErpGMSVLVATAYMEEAE 202
|
|
| PRK10575 |
PRK10575 |
Fe3+-hydroxamate ABC transporter ATP-binding protein FhuC; |
1939-2078 |
4.62e-11 |
|
Fe3+-hydroxamate ABC transporter ATP-binding protein FhuC;
Pssm-ID: 182561 [Multi-domain] Cd Length: 265 Bit Score: 65.58 E-value: 4.62e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1939 PPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQFDAITELLTGREHVEF------ 2011
Cdd:PRK10575 35 PAGKVTGLIGHNGSGKSTLLKMLGRHQPPSEGEILLDAQPLESwSSKAFARKVAYLPQQLPAAEGMTVRELVAIgrypwh 114
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2012 FALLR-GVPEKEvgKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK10575 115 GALGRfGAADRE--KVEE-AISLVGLKPLAHRLVDSLSGGERQRAWIAMLVAQDSRCLLLDEPTSALD 179
|
|
| lolD |
PRK11629 |
lipoprotein-releasing ABC transporter ATP-binding protein LolD; |
1938-2078 |
5.47e-11 |
|
lipoprotein-releasing ABC transporter ATP-binding protein LolD;
Pssm-ID: 183244 [Multi-domain] Cd Length: 233 Bit Score: 64.84 E-value: 5.47e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDAF-----LNKNSILSNIHEVHQNMGYCPQFDAITELLTGREHVEF 2011
Cdd:PRK11629 32 IGEGEMMAIVGSSGSGKSTLLHLLGGlDTP-TSGDVIfngqpMSKLSSAAKAELRNQKLGFIYQFHHLLPDFTALENVAM 110
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2012 FALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK11629 111 PLLIGKKKPAEINSRALEMLAAVGLEHRANHRPSELSGGERQRVAIARALVNNPRLVLADEPTGNLD 177
|
|
| 3a01205 |
TIGR00956 |
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other] |
916-1107 |
5.89e-11 |
|
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273362 [Multi-domain] Cd Length: 1394 Bit Score: 68.21 E-value: 5.89e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLalnFYEGQITSFLGHNGAGKTTTMSILTGlfppTSGTAYILGKDI----RSEMSSIRQNLGVCPQHNVLFDMLTVE 991
Cdd:TIGR00956 782 VDGW---VKPGTLTALMGASGAGKTTLLNVLAE----RVTTGVITGGDRlvngRPLDSSFQRSIGYVQQQDLHLPTSTVR 854
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 992 EHIWFYARLK-----GLSEK--HVKA-----EMEQMAlD--VGLPPSKLKSKtsqlsggmQRK-LSVALAFVGGSKVVI- 1055
Cdd:TIGR00956 855 ESLRFSAYLRqpksvSKSEKmeYVEEvikllEMESYA-DavVGVPGEGLNVE--------QRKrLTIGVELVAKPKLLLf 925
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1056 LDEPTAGVDPYSRRGIWELLLKY-RQGRTiILSTHHMDEADILG--DRIAIISHG 1107
Cdd:TIGR00956 926 LDEPTSGLDSQTAWSICKLMRKLaDHGQA-ILCTIHQPSAILFEefDRLLLLQKG 979
|
|
| PRK14246 |
PRK14246 |
phosphate ABC transporter ATP-binding protein; Provisional |
1928-2143 |
5.92e-11 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172734 [Multi-domain] Cd Length: 257 Bit Score: 65.07 E-value: 5.92e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLT-------GDTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQFDAIT 2000
Cdd:PRK14246 23 KAILKDITIKIPNNSIFGIMGPSGSGKSTLLKVLNrlieiydSKIKVDGKVLYFGKDIFQIDAIKLRKEVGMVFQQPNPF 102
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 ELLTGREHVEFFALLRGVPEK-EVGKVGEWAIRKLGLVKygEKY------ASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:PRK14246 103 PHLSIYDNIAYPLKSHGIKEKrEIKKIVEECLRKVGLWK--EVYdrlnspASQLSGGQQQRLTIARALALKPKVLLMDEP 180
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEgRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL----KNRFGDGYTI 2143
Cdd:PRK14246 181 TSMIDIVNSQAIEKLITELKNE-IAIVIVSHNPQQVARVADYVAFLYNGELVEWGSSNEIftspKNELTEKYVI 253
|
|
| cbiO |
PRK13649 |
energy-coupling factor transporter ATPase; |
1929-2123 |
6.27e-11 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184208 [Multi-domain] Cd Length: 280 Bit Score: 65.54 E-value: 6.27e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTP----VTRGDAFLNKNSILSNIHEVHQNMGYCPQFdAITELL 2003
Cdd:PRK13649 21 RALFDVNLTIEDGSYTAFIGHTGSGKSTIMQLLNGlHVPtqgsVRVDDTLITSTSKNKDIKQIRKKVGLVFQF-PESQLF 99
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2004 --TGREHVEFFALLRGVPEKEVGKVgewAIRKLGLVKYGE----KYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGM 2077
Cdd:PRK13649 100 eeTVLKDVAFGPQNFGVSQEEAEAL---AREKLALVGISEslfeKNPFELSGGQMRRVAIAGILAMEPKILVLDEPTAGL 176
|
170 180 190 200
....*....|....*....|....*....|....*....|....*.
gi 568925948 2078 DPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13649 177 DPKGRKELMTLFKKLHQSGMTIVLVTHLMDDVANYADFVYVLEKGK 222
|
|
| nickel_nikE |
TIGR02769 |
nickel import ATP-binding protein NikE; This family represents the NikE subunit of a ... |
1913-2123 |
6.86e-11 |
|
nickel import ATP-binding protein NikE; This family represents the NikE subunit of a multisubunit nickel import ABC transporter complex. Nickel, once imported, may be used in urease and in certain classes of hydrogenase and superoxide dismutase. [Transport and binding proteins, Cations and iron carrying compounds]
Pssm-ID: 131816 [Multi-domain] Cd Length: 265 Bit Score: 65.21 E-value: 6.86e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYR-------RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAF--------LNKN 1977
Cdd:TIGR02769 2 LLEVRDVTHTYRtgglfgaKQRAPVLTNVSLSIEEGETVGLLGRSGCGKSTLARLLLGLEKPAQGTVSfrgqdlyqLDRK 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1978 SILSNIHEVHQNMGYCPqfDAITELLTGREHV-EFFALLRGVPEKEVGKVGEWAIRKLGL-VKYGEKYASNYSGGNKRKL 2055
Cdd:TIGR02769 82 QRRAFRRDVQLVFQDSP--SAVNPRMTVRQIIgEPLRHLTSLDESEQKARIAELLDMVGLrSEDADKLPRQLSGGQLQRI 159
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2056 STAMALIGGPPVVFLDEPTTGMDpkarRFLWNCALSIVKE-----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:TIGR02769 160 NIARALAVKPKLIVLDEAVSNLD----MVLQAVILELLRKlqqafGTAYLFITHDLRLVQSFCQRVAVMDKGQ 228
|
|
| PRK10575 |
PRK10575 |
Fe3+-hydroxamate ABC transporter ATP-binding protein FhuC; |
888-1118 |
7.01e-11 |
|
Fe3+-hydroxamate ABC transporter ATP-binding protein FhuC;
Pssm-ID: 182561 [Multi-domain] Cd Length: 265 Bit Score: 65.19 E-value: 7.01e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 888 MEEEPTHLRLGVSIQNLVkvYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE 967
Cdd:PRK10575 1 MQEYTNHSDTTFALRNVS--FRVPGRTLLHPLSLTFPAGKVTGLIGHNGSGKSTLLKMLGRHQPPSEGEILLDAQPLESW 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MS-SIRQNLGVCPQHNVLFDMLTVEEHI------WFYARLKGLSEKHVKAEmEQMALdVGLPPSKLKSKTSqLSGGMQRK 1040
Cdd:PRK10575 79 SSkAFARKVAYLPQQLPAAEGMTVRELVaigrypWHGALGRFGAADREKVE-EAISL-VGLKPLAHRLVDS-LSGGERQR 155
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1041 LSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGSSLFL 1118
Cdd:PRK10575 156 AWIAMLVAQDSRCLLLDEPTSALDIAHQVDVLALVhrLSQERGLTVIAVLHDINMAARYCDYLVALRGGEMIAQGTPAEL 235
|
|
| btuD |
PRK09536 |
corrinoid ABC transporter ATPase; Reviewed |
1929-2152 |
9.95e-11 |
|
corrinoid ABC transporter ATPase; Reviewed
Pssm-ID: 236554 [Multi-domain] Cd Length: 402 Bit Score: 66.40 E-value: 9.95e-11
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQFDAITELLTGRE 2007
Cdd:PRK09536 17 TVLDGVDLSVREGSLVGLVGPNGAGKTTLLRAINGTLTPTAGTVLVAGDDVEAlSARAASRRVASVPQDTSLSFEFDVRQ 96
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 HVEFFA---LLRGVPEKEVGK-VGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD-PKAR 2082
Cdd:PRK09536 97 VVEMGRtphRSRFDTWTETDRaAVERAMERTGVAQFADRPVTSLSGGERQRVLLARALAQATPVLLLDEPTASLDiNHQV 176
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2083 RFLwNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG------SVQHLKNRFgDGYTIVvriaGSNP 2152
Cdd:PRK09536 177 RTL-ELVRRLVDDGKTAVAAIHDLDLAARYCDELVLLADGRVRAAGppadvlTADTLRAAF-DARTAV----GTDP 246
|
|
| PRK13546 |
PRK13546 |
teichoic acids export ABC transporter ATP-binding subunit TagH; |
1911-2128 |
1.05e-10 |
|
teichoic acids export ABC transporter ATP-binding subunit TagH;
Pssm-ID: 184131 [Multi-domain] Cd Length: 264 Bit Score: 64.45 E-value: 1.05e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYR--RKRKP----------------AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDa 1972
Cdd:PRK13546 2 NVSVNIKNVTKEYRiyRTNKErmkdalipkhknktffALDDISLKAYEGDVIGLVGINGSGKSTLSNIIGGSLSPTVGK- 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1973 fLNKNSILSNIhevHQNMGYCPQfdaitelLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNK 2052
Cdd:PRK13546 81 -VDRNGEVSVI---AISAGLSGQ-------LTGIENIEFKMLCMGFKRKEIKAMTPKIIEFSELGEFIYQPVKKYSSGMR 149
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2053 RKLSTAMALIGGPPVVFLDEPTTGMDpkaRRFLWNCALSIVK---EGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:PRK13546 150 AKLGFSINITVNPDILVIDEALSVGD---QTFAQKCLDKIYEfkeQNKTIFFVSHNLGQVRQFCTKIAWIEGGKLKDYG 225
|
|
| araG |
PRK11288 |
L-arabinose ABC transporter ATP-binding protein AraG; |
1941-2123 |
1.19e-10 |
|
L-arabinose ABC transporter ATP-binding protein AraG;
Pssm-ID: 183077 [Multi-domain] Cd Length: 501 Bit Score: 66.47 E-value: 1.19e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1941 GECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN--KNSILSNIHEVHQNMGYCPQ---FDAITELLTGRE-------- 2007
Cdd:PRK11288 279 GEIVGLFGLVGAGRSELMKLLYGATRRTAGQVYLDgkPIDIRSPRDAIRAGIMLCPEdrkAEGIIPVHSVADninisarr 358
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 -HVEFFALLRGVPEKEVGkvgEWAIRKLGlVK--YGEKYASNYSGGNKRK------LSTAMAliggppVVFLDEPTTGMD 2078
Cdd:PRK11288 359 hHLRAGCLINNRWEAENA---DRFIRSLN-IKtpSREQLIMNLSGGNQQKailgrwLSEDMK------VILLDEPTRGID 428
|
170 180 190 200
....*....|....*....|....*....|....*....|....*
gi 568925948 2079 PKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK11288 429 VGAKHEIYNVIYELAAQGVAVLFVSSDLPEVLGVADRIVVMREGR 473
|
|
| YejF |
COG4172 |
ABC-type microcin C transport system, duplicated ATPase component YejF [Secondary metabolites ... |
900-1109 |
1.22e-10 |
|
ABC-type microcin C transport system, duplicated ATPase component YejF [Secondary metabolites biosynthesis, transport and catabolism];
Pssm-ID: 443332 [Multi-domain] Cd Length: 533 Bit Score: 66.63 E-value: 1.22e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYR--DGMKVAVDGLALNFYEGQITSFLGHNGAGKT-TTMSILtGLFPP----TSGTAYILGKDI----RSEM 968
Cdd:COG4172 8 SVEDLSVAFGqgGGTVEAVKGVSFDIAAGETLALVGESGSGKSvTALSIL-RLLPDpaahPSGSILFDGQDLlglsEREL 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 SSIRQN------------LgvcpqhNVLFdmlTVEEHIWFYARL-KGLSEKHVKAEMEQMALDVGLPP--SKLKSKTSQL 1033
Cdd:COG4172 87 RRIRGNriamifqepmtsL------NPLH---TIGKQIAEVLRLhRGLSGAAARARALELLERVGIPDpeRRLDAYPHQL 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1034 SGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHhmDeadiLG------DRIAIIS 1105
Cdd:COG4172 158 SGGQRQRVMIAMALANEPDLLIADEPTTALDVTVQAQILDLLkdLQRELGMALLLITH--D----LGvvrrfaDRVAVMR 231
|
....
gi 568925948 1106 HGKL 1109
Cdd:COG4172 232 QGEI 235
|
|
| PRK13539 |
PRK13539 |
cytochrome c biogenesis protein CcmA; Provisional |
1940-2105 |
1.26e-10 |
|
cytochrome c biogenesis protein CcmA; Provisional
Pssm-ID: 237421 [Multi-domain] Cd Length: 207 Bit Score: 63.35 E-value: 1.26e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1940 PGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN-KNSILSNIHEV-----HQNmgycpqfdAITELLTGREHVEFFA 2013
Cdd:PRK13539 27 AGEALVLTGPNGSGKTTLLRLIAGLLPPAAGTIKLDgGDIDDPDVAEAchylgHRN--------AMKPALTVAENLEFWA 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2014 LLRGVPEKEVgkvgEWAIRKLGL-----VKYGekyasNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNC 2088
Cdd:PRK13539 99 AFLGGEELDI----AAALEAVGLaplahLPFG-----YLSAGQKRRVALARLLVSNRPIWILDEPTAALDAAAVALFAEL 169
|
170
....*....|....*..
gi 568925948 2089 ALSIVKEGRSVVLTSHS 2105
Cdd:PRK13539 170 IRAHLAQGGIVIAATHI 186
|
|
| PLN03140 |
PLN03140 |
ABC transporter G family member; Provisional |
1940-2104 |
1.40e-10 |
|
ABC transporter G family member; Provisional
Pssm-ID: 215599 [Multi-domain] Cd Length: 1470 Bit Score: 67.18 E-value: 1.40e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1940 PGECFGLLGVNGAGKSTTFKMLTGDTP--VTRGD----AFLNKNSILSNIHevhqnmGYCPQFDAITELLTGREHVEFFA 2013
Cdd:PLN03140 905 PGVLTALMGVSGAGKTTLMDVLAGRKTggYIEGDirisGFPKKQETFARIS------GYCEQNDIHSPQVTVRESLIYSA 978
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2014 LLRgVPeKEVGK---------VGEWA-IRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARR 2083
Cdd:PLN03140 979 FLR-LP-KEVSKeekmmfvdeVMELVeLDNLKDAIVGLPGVTGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAA 1056
|
170 180
....*....|....*....|.
gi 568925948 2084 FLWNCALSIVKEGRSVVLTSH 2104
Cdd:PLN03140 1057 IVMRTVRNTVDTGRTVVCTIH 1077
|
|
| PRK11264 |
PRK11264 |
putative amino-acid ABC transporter ATP-binding protein YecC; Provisional |
1914-2123 |
1.40e-10 |
|
putative amino-acid ABC transporter ATP-binding protein YecC; Provisional
Pssm-ID: 183063 [Multi-domain] Cd Length: 250 Bit Score: 64.00 E-value: 1.40e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTP----VTRGDAFLNKNSILSN----IH 1984
Cdd:PRK11264 4 IEVKNLVKKF--HGQTVLHGIDLEVKPGEVVAIIGPSGSGKTTLLRCINLlEQPeagtIRVGDITIDTARSLSQqkglIR 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVHQNMGYCPQFDAITELLTGREHV-EFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIG 2063
Cdd:PRK11264 82 QLRQHVGFVFQNFNLFPHRTVLENIiEGPVIVKGEPKEEATARARELLAKVGLAGKETSYPRRLSGGQQQRVAIARALAM 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2064 GPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK11264 162 RPEVILFDEPTSALDPELVGEVLNTIRQLAQEKRTMVIVTHEMSFARDVADRAIFMDQGR 221
|
|
| fbpC |
PRK11432 |
ferric ABC transporter ATP-binding protein; |
1910-2083 |
1.40e-10 |
|
ferric ABC transporter ATP-binding protein;
Pssm-ID: 183133 [Multi-domain] Cd Length: 351 Bit Score: 65.51 E-value: 1.40e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTKiyRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN-----KNSIlsnih 1984
Cdd:PRK11432 3 QKNFVVLKNITK--RFGSNTVIDNLNLTIKQGTMVTLLGPSGCGKTTVLRLVAGLEKPTEGQIFIDgedvtHRSI----- 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 evhQNMGYCPQFD--AITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALI 2062
Cdd:PRK11432 76 ---QQRDICMVFQsyALFPHMSLGENVGYGLKMLGVPKEERKQRVKEALELVDLAGFEDRYVDQISGGQQQRVALARALI 152
|
170 180
....*....|....*....|.
gi 568925948 2063 GGPPVVFLDEPTTGMDPKARR 2083
Cdd:PRK11432 153 LKPKVLLFDEPLSNLDANLRR 173
|
|
| xylG |
TIGR02633 |
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose ... |
1913-2123 |
1.50e-10 |
|
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose isomerase and xylulokinase enzymes for xylose utilization. Members of this protein family are the ATP-binding cassette (ABC) subunit of the known or predicted high-affinity xylose ABC transporter for xylose import. These genes, which closely resemble other sugar transport ABC transporter genes, typically are encoded near xylose utilization enzymes and regulatory proteins. Note that this form of the transporter contains two copies of the ABC transporter domain (pfam00005). [Transport and binding proteins, Carbohydrates, organic alcohols, and acids]
Pssm-ID: 131681 [Multi-domain] Cd Length: 500 Bit Score: 66.00 E-value: 1.50e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP--VTRGDAFLNKNSIL-SNIHE---- 1985
Cdd:TIGR02633 1 LLEMKGIVKTFGGVK--ALDGIDLEVRPGECVGLCGENGAGKSTLMKILSGVYPhgTWDGEIYWSGSPLKaSNIRDtera 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 ----VHQNMGYCPQFDAITELLTGREhveffALLRG--VPEKEVGKVGEWAIRKLGL-VKYGEKYASNYSGGNKRKLSTA 2058
Cdd:TIGR02633 79 giviIHQELTLVPELSVAENIFLGNE-----ITLPGgrMAYNAMYLRAKNLLRELQLdADNVTRPVGDYGGGQQQLVEIA 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2059 MALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:TIGR02633 154 KALNKQARLLILDEPSSSLTEKETEILLDIIRDLKAHGVACVYISHKLNEVKAVCDTICVIRDGQ 218
|
|
| ABCC_NFT1 |
cd03369 |
ATP-binding cassette domain 2 of NFT1, subfamily C; Domain 2 of NFT1 (New full-length MRP-type ... |
892-1109 |
1.91e-10 |
|
ATP-binding cassette domain 2 of NFT1, subfamily C; Domain 2 of NFT1 (New full-length MRP-type transporter 1). NFT1 belongs to the MRP (multidrug resistance-associated protein) family of ABC transporters. Some of the MRP members have five additional transmembrane segments in their N-terminus, but the function of these additional membrane-spanning domains is not clear. The MRP was found in the multidrug-resisting lung cancer cell in which p-glycoprotein was not overexpressed. MRP exports glutathione by drug stimulation, as well as, certain substrates in conjugated forms with anions such as glutathione, glucuronate, and sulfate.
Pssm-ID: 213269 [Multi-domain] Cd Length: 207 Bit Score: 62.81 E-value: 1.91e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 892 PTHLRlgVSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSS 970
Cdd:cd03369 2 PEHGE--IEVENLSVRYAPDLPPVLKNVSFKVKAGEKIGIVGRTGAGKSTLILALFRFLEAEEGKIEIDGIDISTiPLED 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 971 IRQNLGVCPQHNVLFdMLTVEEHIWFYARlkgLSEKHVKAemeqmALDVglppsklKSKTSQLSGGMQRKLSVALAFVGG 1050
Cdd:cd03369 80 LRSSLTIIPQDPTLF-SGTIRSNLDPFDE---YSDEEIYG-----ALRV-------SEGGLNLSQGQRQLLCLARALLKR 143
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1051 SKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDE-ADIlgDRIAIISHGKL 1109
Cdd:cd03369 144 PRVLVLDEATASIDYATDALIQKTIREEFTNSTILTIAHRLRTiIDY--DKILVMDAGEV 201
|
|
| modC_ABC |
TIGR02142 |
molybdenum ABC transporter, ATP-binding protein; This model represents the ATP-binding ... |
928-1114 |
1.93e-10 |
|
molybdenum ABC transporter, ATP-binding protein; This model represents the ATP-binding cassette (ABC) protein of the three subunit molybdate ABC transporter. The three proteins of this complex are homologous to proteins of the sulfate ABC transporter. Molybdenum may be used in nitrogenases of nitrogen-fixing bacteria and in molybdopterin cofactors. In some cases, molybdate may be transported by a sulfate transporter rather than by a specific molybdate transporter. [Transport and binding proteins, Anions]
Pssm-ID: 131197 [Multi-domain] Cd Length: 354 Bit Score: 65.13 E-value: 1.93e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 928 ITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSI-----RQNLGVCPQHNVLFDMLTVEEHIWF-YARLK 1001
Cdd:TIGR02142 25 VTAIFGRSGSGKTTLIRLIAGLTRPDEGEIVLNGRTLFDSRKGIflppeKRRIGYVFQEARLFPHLSVRGNLRYgMKRAR 104
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1002 GLSEKHVKAEMEQMaldVGLPPSkLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ- 1080
Cdd:TIGR02142 105 PSERRISFERVIEL---LGIGHL-LGRLPGRLSGGEKQRVAIGRALLSSPRLLLMDEPLAALDDPRKYEILPYLERLHAe 180
|
170 180 190
....*....|....*....|....*....|....*
gi 568925948 1081 -GRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:TIGR02142 181 fGIPILYVSHSLQEVLRLADRVVVLEDGRVAAAGP 215
|
|
| ABCF_EF-3 |
cd03221 |
ATP-binding cassette domain of elongation factor 3, subfamily F; Elongation factor 3 (EF-3) is ... |
1914-2104 |
2.46e-10 |
|
ATP-binding cassette domain of elongation factor 3, subfamily F; Elongation factor 3 (EF-3) is a cytosolic protein required by fungal ribosomes for in vitro protein synthesis and for in vivo growth. EF-3 stimulates the binding of the EF-1: GTP: aa-tRNA ternary complex to the ribosomal A site by facilitated release of the deacylated tRNA from the E site. The reaction requires ATP hydrolysis. EF-3 contains two ATP nucleotide binding sequence (NBS) motifs. NBSI is sufficient for the intrinsic ATPase activity. NBSII is essential for the ribosome-stimulated functions.
Pssm-ID: 213188 [Multi-domain] Cd Length: 144 Bit Score: 60.54 E-value: 2.46e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilsnihevhQNMGYC 1993
Cdd:cd03221 1 IELENLSKTY--GGKLLLKDISLTINPGDRIGLVGRNGAGKSTLLKLIAGELEPDEGIVTWGST----------VKIGYF 68
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFdaitelltgrehveffallrgvpekevgkvgewairklglvkygekyasnySGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:cd03221 69 EQL---------------------------------------------------SGGEKMRLALAKLLLENPNLLLLDEP 97
|
170 180 190
....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNcalSIVKEGRSVVLTSH 2104
Cdd:cd03221 98 TNHLDLESIEALEE---ALKEYPGTVILVSH 125
|
|
| PRK11176 |
PRK11176 |
lipid A ABC transporter ATP-binding protein/permease MsbA; |
915-1123 |
2.47e-10 |
|
lipid A ABC transporter ATP-binding protein/permease MsbA;
Pssm-ID: 183016 [Multi-domain] Cd Length: 582 Bit Score: 65.81 E-value: 2.47e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLFDMlTVEEH 993
Cdd:PRK11176 358 ALRNINFKIPAGKTVALVGRSGSGKSTIANLLTRFYDIDEGEILLDGHDLRDyTLASLRNQVALVSQNVHLFND-TIANN 436
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 994 IwFYARlkglSEKHVKAEMEQMA-----------LDVGLPPSKLKSKTSqLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:PRK11176 437 I-AYAR----TEQYSREQIEEAArmayamdfinkMDNGLDTVIGENGVL-LSGGQRQRIAIARALLRDSPILILDEATSA 510
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 1063 VDPYSRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGSSLFLKNQLG 1123
Cdd:PRK11176 511 LDTESERAIQAALDELQKNRTSLVIAHRLSTIE-KADEILVVEDGEIVERGTHAELLAQNG 570
|
|
| PRK15439 |
PRK15439 |
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional |
919-1109 |
4.47e-10 |
|
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional
Pssm-ID: 185336 [Multi-domain] Cd Length: 510 Bit Score: 64.69 E-value: 4.47e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 919 LALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVC------PQHNVLFDM----- 987
Cdd:PRK15439 282 ISLEVRAGEILGLAGVVGAGRTELAETLYGLRPARGGRIMLNGKEINALSTAQRLARGLVylpedrQSSGLYLDAplawn 361
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 988 ---LTVEEHIWFyarlkgLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVD 1064
Cdd:PRK15439 362 vcaLTHNRRGFW------IKPARENAVLERYRRALNIKFNHAEQAARTLSGGNQQKVLIAKCLEASPQLLIVDEPTRGVD 435
|
170 180 190 200
....*....|....*....|....*....|....*....|....*.
gi 568925948 1065 PYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK15439 436 VSARNDIYQLIRSIaAQNVAVLFISSDLEEIEQMADRVLVMHQGEI 481
|
|
| cbiO |
PRK13634 |
cobalt transporter ATP-binding subunit; Provisional |
1938-2133 |
5.90e-10 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237454 [Multi-domain] Cd Length: 290 Bit Score: 62.73 E-value: 5.90e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-----NIHEVHQNMGYCPQFdaiTELLTGREHVE-- 2010
Cdd:PRK13634 30 IPSGSYVAIIGHTGSGKSTLLQHLNGLLQPTSGTVTIGERVITAgkknkKLKPLRKKVGIVFQF---PEHQLFEETVEkd 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2011 --FFALLRGVPEKEV-GKVGEWairkLGLVKYGEKYAS----NYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARR 2083
Cdd:PRK13634 107 icFGPMNFGVSEEDAkQKAREM----IELVGLPEELLArspfELSGGQMRRVAIAGVLAMEPEVLVLDEPTAGLDPKGRK 182
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2084 FLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK13634 183 EMMEMFYKLHKEkGLTTVLVTHSMEDAARYADQIVVMHKGTVFLQGTPREI 233
|
|
| ABCC_Hemolysin |
cd03252 |
ATP-binding cassette domain of hemolysin B, subfamily C; The ABC-transporter hemolysin B is a ... |
1923-2138 |
6.05e-10 |
|
ATP-binding cassette domain of hemolysin B, subfamily C; The ABC-transporter hemolysin B is a central component of the secretion machinery that translocates the toxin, hemolysin A, in a Sec-independent fashion across both membranes of E. coli. The hemolysin A (HlyA) transport machinery is composed of the ATP-binding cassette (ABC) transporter HlyB located in the inner membrane, hemolysin D (HlyD), also anchored in the inner membrane, and TolC, which resides in the outer membrane. HlyD apparently forms a continuous channel that bridges the entire periplasm, interacting with TolC and HlyB. This arrangement prevents the appearance of periplasmic intermediates of HlyA during substrate transport. Little is known about the molecular details of HlyA transport, but it is evident that ATP-hydrolysis by the ABC-transporter HlyB is a necessary source of energy.
Pssm-ID: 213219 [Multi-domain] Cd Length: 237 Bit Score: 61.73 E-value: 6.05e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1923 YRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHEVHQNMGYCPQfdaiTE 2001
Cdd:cd03252 10 YKPDGPVILDNISLRIKPGEVVGIVGRSGSGKSTLTKLIQRFYVPENGRVLVDGHDLaLADPAWLRRQVGVVLQ----EN 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2002 LLTGREHVEFFALLR-GVPEKEVGKVGEWA-----IRKLGL---VKYGEKYASnYSGGNKRKLSTAMALIGGPPVVFLDE 2072
Cdd:cd03252 86 VLFNRSIRDNIALADpGMSMERVIEAAKLAgahdfISELPEgydTIVGEQGAG-LSGGQRQRIAIARALIHNPRILIFDE 164
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2073 PTTGMDPKARRFLWNCALSIVKeGRSVVLTSHSMEECEAlCTRMAIMVNGRFRCLGSVQHLKNRFG 2138
Cdd:cd03252 165 ATSALDYESEHAIMRNMHDICA-GRTVIIIAHRLSTVKN-ADRIIVMEKGRIVEQGSHDELLAENG 228
|
|
| PRK15439 |
PRK15439 |
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional |
1934-2124 |
6.42e-10 |
|
autoinducer 2 ABC transporter ATP-binding protein LsrA; Provisional
Pssm-ID: 185336 [Multi-domain] Cd Length: 510 Bit Score: 64.30 E-value: 6.42e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1934 ICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI--LSNIHEVHQNMGYCPQ--------FDA----- 1998
Cdd:PRK15439 282 ISLEVRAGEILGLAGVVGAGRTELAETLYGLRPARGGRIMLNGKEInaLSTAQRLARGLVYLPEdrqssglyLDAplawn 361
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1999 ITELLTGRehVEFFalLRgvPEKEvGKVGEWAIRKLGL-VKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGM 2077
Cdd:PRK15439 362 VCALTHNR--RGFW--IK--PARE-NAVLERYRRALNIkFNHAEQAARTLSGGNQQKVLIAKCLEASPQLLIVDEPTRGV 434
|
170 180 190 200
....*....|....*....|....*....|....*....|....*..
gi 568925948 2078 DPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:PRK15439 435 DVSARNDIYQLIRSIAAQNVAVLFISSDLEEIEQMADRVLVMHQGEI 481
|
|
| PRK10762 |
PRK10762 |
D-ribose transporter ATP binding protein; Provisional |
1914-2123 |
6.45e-10 |
|
D-ribose transporter ATP binding protein; Provisional
Pssm-ID: 236755 [Multi-domain] Cd Length: 501 Bit Score: 64.25 E-value: 6.45e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKiyrrkrkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSN----------- 1982
Cdd:PRK10762 258 LKVDNLSG-------PGVNDVSFTLRKGEILGVSGLMGAGRTELMKVLYGALPRTSGYVTLDGHEVVTRspqdglangiv 330
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1983 -IHE------------VHQNMGYCpqfdAITEL--LTGRehveffalLRGVPEKEVgkVGEWaIRkLGLVKYG--EKYAS 2045
Cdd:PRK10762 331 yISEdrkrdglvlgmsVKENMSLT----ALRYFsrAGGS--------LKHADEQQA--VSDF-IR-LFNIKTPsmEQAIG 394
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2046 NYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK10762 395 LLSGGNQQKVAIARGLMTRPKVLILDEPTRGVDVGAKKEIYQLINQFKAEGLSIILVSSEMPEVLGMSDRILVMHEGR 472
|
|
| PRK15134 |
PRK15134 |
microcin C ABC transporter ATP-binding protein YejF; Provisional |
900-1112 |
6.50e-10 |
|
microcin C ABC transporter ATP-binding protein YejF; Provisional
Pssm-ID: 237917 [Multi-domain] Cd Length: 529 Bit Score: 64.34 E-value: 6.50e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 900 SIQNLVKVYRDGMKV--AVDGLALNFYEGQITSFLGHNGAGKT-TTMSILTGLfpPTSGTAYILGkDIR----------- 965
Cdd:PRK15134 7 AIENLSVAFRQQQTVrtVVNDVSLQIEAGETLALVGESGSGKSvTALSILRLL--PSPPVVYPSG-DIRfhgesllhase 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 966 SEMSSIRQN----------LGVCPQHNV---LFDMLTVEehiwfyarlKGLSEKHVKAEMEQMALDVGL--PPSKLKSKT 1030
Cdd:PRK15134 84 QTLRGVRGNkiamifqepmVSLNPLHTLekqLYEVLSLH---------RGMRREAARGEILNCLDRVGIrqAAKRLTDYP 154
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1031 SQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:PRK15134 155 HQLSGGERQRVMIAMALLTRPELLIADEPTTALDVSVQAQILQLLreLQQELNMGLLFITHNLSIVRKLADRVAVMQNGR 234
|
....
gi 568925948 1109 lcCV 1112
Cdd:PRK15134 235 --CV 236
|
|
| ABCC_TAP |
cd03248 |
ATP-binding cassette domain of the Transporter Associated with Antigen Processing, subfamily C; ... |
1913-2110 |
6.98e-10 |
|
ATP-binding cassette domain of the Transporter Associated with Antigen Processing, subfamily C; TAP (Transporter Associated with Antigen Processing) is essential for peptide delivery from the cytosol into the lumen of the endoplasmic reticulum (ER), where these peptides are loaded on major histocompatibility complex (MHC) I molecules. Loaded MHC I leave the ER and display their antigenic cargo on the cell surface to cytotoxic T cells. Subsequently, virus-infected or malignantly transformed cells can be eliminated. TAP belongs to the large family of ATP-binding cassette (ABC) transporters, which translocate a vast variety of solutes across membranes.
Pssm-ID: 213215 [Multi-domain] Cd Length: 226 Bit Score: 61.33 E-value: 6.98e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYR-RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHE-VHQNM 1990
Cdd:cd03248 11 IVKFQNVTFAYPtRPDTLVLQDVSFTLHPGEVTALVGPSGSGKSTVVALLENFYQPQGGQVLLDGKPISQYEHKyLHSKV 90
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 GycpqfdaitelLTGREHVEFFALLR-----GVPEKEVGKVGEWA--------IRKLGLVKY---GEKyASNYSGGNKRK 2054
Cdd:cd03248 91 S-----------LVGQEPVLFARSLQdniayGLQSCSFECVKEAAqkahahsfISELASGYDtevGEK-GSQLSGGQKQR 158
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2055 LSTAMALIGGPPVVFLDEPTTGMDPKARRFLwNCALSIVKEGRSVVLTSHSMEECE 2110
Cdd:cd03248 159 VAIARALIRNPQVLILDEATSALDAESEQQV-QQALYDWPERRTVLVIAHRLSTVE 213
|
|
| ArpD |
COG4618 |
ABC-type protease/lipase transport system, ATPase and permease components [Intracellular ... |
1927-2128 |
8.17e-10 |
|
ABC-type protease/lipase transport system, ATPase and permease components [Intracellular trafficking, secretion, and vesicular transport];
Pssm-ID: 443660 [Multi-domain] Cd Length: 563 Bit Score: 64.00 E-value: 8.17e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1927 RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI-LSNIHEVHQNMGYCPQfdaitelltg 2005
Cdd:COG4618 344 KRPILRGVSFSLEPGEVLGVIGPSGSGKSTLARLLVGVWPPTAGSVRLDGADLsQWDREELGRHIGYLPQ---------- 413
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2006 reHVEFF--------ALLRGV-PEKevgkVGEWA--------IRKL--GlvkY----GEKyASNYSGGNKRKLSTAMALI 2062
Cdd:COG4618 414 --DVELFdgtiaeniARFGDAdPEK----VVAAAklagvhemILRLpdG---YdtriGEG-GARLSGGQRQRIGLARALY 483
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2063 GGPPVVFLDEPTTGMDPKARRFLwNCALSIVKE-GRSVVLTSHSMeecEAL--CTRMAIMVNGRFRCLG 2128
Cdd:COG4618 484 GDPRLVVLDEPNSNLDDEGEAAL-AAAIRALKArGATVVVITHRP---SLLaaVDKLLVLRDGRVQAFG 548
|
|
| cbiO |
PRK13642 |
energy-coupling factor transporter ATPase; |
1911-2122 |
9.65e-10 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184202 [Multi-domain] Cd Length: 277 Bit Score: 62.03 E-value: 9.65e-10
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELtkIYRRKRKPAVDR---ICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEV 1986
Cdd:PRK13642 2 NKILEVENL--VFKYEKESDVNQlngVSFSITKGEWVSIIGQNGSGKSTTARLIDGLFEEFEGKVKIDGELLTAeNVWNL 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNMGYCPQF-DAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:PRK13642 80 RRKIGMVFQNpDNQFVGATVEDDVAFGMENQGIPREEMIKRVDEALLAVNMLDFKTREPARLSGGQKQRVAVAGIIALRP 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLT-SHSMEECeALCTRMAIMVNG 2122
Cdd:PRK13642 160 EIIILDESTSMLDPTGRQEIMRVIHEIKEKYQLTVLSiTHDLDEA-ASSDRILVMKAG 216
|
|
| PRK10522 |
PRK10522 |
multidrug transporter membrane component/ATP-binding component; Provisional |
899-1110 |
1.05e-09 |
|
multidrug transporter membrane component/ATP-binding component; Provisional
Pssm-ID: 236707 [Multi-domain] Cd Length: 547 Bit Score: 63.45 E-value: 1.05e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQnlgv 977
Cdd:PRK10522 323 LELRNVTFAYQD-NGFSVGPINLTIKRGELLFLIGGNGSGKSTLAMLLTGLYQPQSGEILLDGKPVTAEqPEDYRK---- 397
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 cpqhnvLFDmlTVEEHIWFYARLKGLSEKHVKAEmeqmALDVGLPPSKLKSKTS---------QLSGGMQRKLSVALAFV 1048
Cdd:PRK10522 398 ------LFS--AVFTDFHLFDQLLGPEGKPANPA----LVEKWLERLKMAHKLEledgrisnlKLSKGQKKRLALLLALA 465
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHmDEADILGDRIAIISHGKLC 1110
Cdd:PRK10522 466 EERDILLLDEWAADQDPHFRREFYQVLLPLLQemGKTIFAISHD-DHYFIHADRLLEMRNGQLS 528
|
|
| met_CoM_red_A2 |
TIGR03269 |
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in ... |
1914-2137 |
1.08e-09 |
|
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in methanogenesis, methyl coenzyme M reductase, contains alpha, beta, and gamma chains. In older literature, the complex of alpha, beta, and gamma chains was termed component C, while this single chain protein was termed methyl coenzyme M reductase system component A2. [Energy metabolism, Methanogenesis]
Pssm-ID: 132313 [Multi-domain] Cd Length: 520 Bit Score: 63.67 E-value: 1.08e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG--DTPVTRGdaflnknsilsnihEVHQNMG 1991
Cdd:TIGR03269 1 IEVKNLTKKFDGKE--VLKNISFTIEEGEVLGILGRSGAGKSVLMHVLRGmdQYEPTSG--------------RIIYHVA 64
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQF-------------------------------DAITELLTGREHVEF---FAL------LRGVPE--KEVGKVGEW 2029
Cdd:TIGR03269 65 LCEKCgyverpskvgepcpvcggtlepeevdfwnlsDKLRRRIRKRIAIMLqrtFALygddtvLDNVLEalEEIGYEGKE 144
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2030 AIRK----LGLVKYGEKY---ASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVL 2101
Cdd:TIGR03269 145 AVGRavdlIEMVQLSHRIthiARDLSGGEKQRVVLARQLAKEPFLFLADEPTGTLDPQTAKLVHNALEEAVKAsGISMVL 224
|
250 260 270
....*....|....*....|....*....|....*.
gi 568925948 2102 TSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRF 2137
Cdd:TIGR03269 225 TSHWPEVIEDLSDKAIWLENGEIKEEGTPDEVVAVF 260
|
|
| ABCC_MRP_domain1 |
cd03250 |
ATP-binding cassette domain 1 of multidrug resistance-associated protein, subfamily C; This ... |
1927-2123 |
1.09e-09 |
|
ATP-binding cassette domain 1 of multidrug resistance-associated protein, subfamily C; This subfamily is also known as MRP (multidrug resistance-associated protein). Some of the MRP members have five additional transmembrane segments in their N-terminus, but the function of these additional membrane-spanning domains is not clear. The MRP was found in the multidrug-resisting lung cancer cell in which p-glycoprotein was not overexpressed. MRP exports glutathione by drug stimulation, as well as, certain substrates in conjugated forms with anions, such as glutathione, glucuronate, and sulfate.
Pssm-ID: 213217 [Multi-domain] Cd Length: 204 Bit Score: 60.18 E-value: 1.09e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1927 RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilsnihevhqnMGYCPQFDAItelLTG- 2005
Cdd:cd03250 17 TSFTLKDINLEVPKGELVAIVGPVGSGKSSLLSALLGELEKLSGSVSVPGS------------IAYVSQEPWI---QNGt 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2006 -REHVEFfallrGVPEKEvgkvgEW---AIRKLGLVK------------YGEKyASNYSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:cd03250 82 iRENILF-----GKPFDE-----ERyekVIKACALEPdleilpdgdlteIGEK-GINLSGGQKQRISLARAVYSDADIYL 150
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2070 LDEPTTGMDPKARRFLW-NCALSIVKEGRSVVLTSHSMEECEAlCTRMAIMVNGR 2123
Cdd:cd03250 151 LDDPLSAVDAHVGRHIFeNCILGLLLNNKTRILVTHQLQLLPH-ADQIVVLDNGR 204
|
|
| cbiO |
PRK13638 |
energy-coupling factor ABC transporter ATP-binding protein; |
1923-2107 |
1.16e-09 |
|
energy-coupling factor ABC transporter ATP-binding protein;
Pssm-ID: 184198 [Multi-domain] Cd Length: 271 Bit Score: 61.56 E-value: 1.16e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1923 YRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAF-------LNKNSILSniheVHQNMGYCPQ 1995
Cdd:PRK13638 9 FRYQDEPVLKGLNLDFSLSPVTGLVGANGCGKSTLFMNLSGLLRPQKGAVLwqgkpldYSKRGLLA----LRQQVATVFQ 84
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1996 FDAITELLTGREHVEFFALLR-GVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPT 2074
Cdd:PRK13638 85 DPEQQIFYTDIDSDIAFSLRNlGVPEAEITRRVDEALTLVDAQHFRHQPIQCLSHGQKKRVAIAGALVLQARYLLLDEPT 164
|
170 180 190
....*....|....*....|....*....|...
gi 568925948 2075 TGMDPKARRFLWNCALSIVKEGRSVVLTSHSME 2107
Cdd:PRK13638 165 AGLDPAGRTQMIAIIRRIVAQGNHVIISSHDID 197
|
|
| PvdE |
COG4615 |
ABC-type siderophore export system, fused ATPase and permease components [Inorganic ion ... |
921-1089 |
1.17e-09 |
|
ABC-type siderophore export system, fused ATPase and permease components [Inorganic ion transport and metabolism];
Pssm-ID: 443659 [Multi-domain] Cd Length: 547 Bit Score: 63.28 E-value: 1.17e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFYEGQITsFL-GHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGV--CPQHnvLFDMLtveehiwf 996
Cdd:COG4615 353 LTIRRGELV-FIvGGNGSGKSTLAKLLTGLYRPESGEILLDGQPVTADnREAYRQLFSAvfSDFH--LFDRL-------- 421
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 997 YARLKGLSEKHVKAEMEQMALDvglppSKLKSK-----TSQLSGGmQRK-LSVALAFVGGSKVVILDEPTAGVDPYSRRg 1070
Cdd:COG4615 422 LGLDGEADPARARELLERLELD-----HKVSVEdgrfsTTDLSQG-QRKrLALLVALLEDRPILVFDEWAADQDPEFRR- 494
|
170 180
....*....|....*....|....
gi 568925948 1071 iW---ELL--LKyRQGRTIILSTH 1089
Cdd:COG4615 495 -VfytELLpeLK-ARGKTVIAISH 516
|
|
| ABC_ThiQ_thiamine_transporter |
cd03298 |
ATP-binding cassette domain of the thiamine transport system; Part of the ... |
1918-2123 |
1.33e-09 |
|
ATP-binding cassette domain of the thiamine transport system; Part of the binding-protein-dependent transport system tbpA-thiPQ for thiamine and TPP. Probably responsible for the translocation of thiamine across the membrane. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213265 [Multi-domain] Cd Length: 211 Bit Score: 60.20 E-value: 1.33e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1918 ELTKIYRRKRKPAVDRICIgIPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDAFLN-------------------KN 1977
Cdd:cd03298 2 RLDKIRFSYGEQPMHFDLT-FAQGEITAIVGPSGSGKSTLLNLIAGfETP-QSGRVLINgvdvtaappadrpvsmlfqEN 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1978 SILSNIhEVHQNMGYCpqfdaitelLTGREHveffalLRGVPEKEVGKvgewAIRKLGLVKYGEKYASNYSGGNKRKLST 2057
Cdd:cd03298 80 NLFAHL-TVEQNVGLG---------LSPGLK------LTAEDRQAIEV----ALARVGLAGLEKRLPGELSGGERQRVAL 139
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:cd03298 140 ARVLVRDKPVLLLDEPFAALDPALRAEMLDLVLDLHAEtKMTVLMVTHQPEDAKRLAQRVVFLDNGR 206
|
|
| ABC_FeS_Assembly |
cd03217 |
ABC-type transport system involved in Fe-S cluster assembly, ATPase component; Biosynthesis of ... |
1914-2130 |
1.58e-09 |
|
ABC-type transport system involved in Fe-S cluster assembly, ATPase component; Biosynthesis of iron-sulfur clusters (Fe-S) depends on multi-protein systems. The SUF system of E. coli and Erwinia chrysanthemi is important for Fe-S biogenesis under stressful conditions. The SUF system is made of six proteins: SufC is an atypical cytoplasmic ABC-ATPase, which forms a complex with SufB and SufD; SufA plays the role of a scaffold protein for assembly of iron-sulfur clusters and delivery to target proteins; SufS is a cysteine desulfurase which mobilizes the sulfur atom from cysteine and provides it to the cluster; SufE has no associated function yet.
Pssm-ID: 213184 [Multi-domain] Cd Length: 200 Bit Score: 59.85 E-value: 1.58e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG--DTPVTRGDAFLNKNSILS-NIHEvHQNM 1990
Cdd:cd03217 1 LEIKDLH--VSVGGKEILKGVNLTIKKGEVHALMGPNGSGKSTLAKTIMGhpKYEVTEGEILFKGEDITDlPPEE-RARL 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1991 G------YCPQFDAITelltgrehVEFFalLRGVPEkevgkvgewairklglvkygekyasNYSGGNKRKLSTAMALIGG 2064
Cdd:cd03217 78 GiflafqYPPEIPGVK--------NADF--LRYVNE-------------------------GFSGGEKKRNEILQLLLLE 122
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2065 PPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEAL-CTRMAIMVNGRFRCLGSV 2130
Cdd:cd03217 123 PDLAILDEPDSGLDIDALRLVAEVINKLREEGKSVLIITHYQRLLDYIkPDRVHVLYDGRIVKSGDK 189
|
|
| PhnL |
COG4778 |
Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [Inorganic ion ... |
901-1107 |
1.78e-09 |
|
Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [Inorganic ion transport and metabolism];
Pssm-ID: 443809 [Multi-domain] Cd Length: 229 Bit Score: 60.14 E-value: 1.78e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKVY----RDGMK-VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayILgkdIRS--------- 966
Cdd:COG4778 7 VENLSKTFtlhlQGGKRlPVLDGVSFSVAAGECVALTGPSGAGKSTLLKCIYGNYLPDSGS--IL---VRHdggwvdlaq 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 967 ----EMSSIRQN-LGVCPQH-NVL-----FDmlTVEEHiwfyARLKGLSEKHVKAEMEQM--ALDV-----GLPPSklks 1028
Cdd:COG4778 82 asprEILALRRRtIGYVSQFlRVIprvsaLD--VVAEP----LLERGVDREEARARARELlaRLNLperlwDLPPA---- 151
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1029 kTsqLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEADILGDRIAIISHG 1107
Cdd:COG4778 152 -T--FSGGEQQRVNIARGFIADPPLLLLDEPTASLDAANRAVVVELIEEAkARGTAIIGIFHDEEVREAVADRVVDVTPF 228
|
|
| PLN03232 |
PLN03232 |
ABC transporter C family member; Provisional |
921-1107 |
1.98e-09 |
|
ABC transporter C family member; Provisional
Pssm-ID: 215640 [Multi-domain] Cd Length: 1495 Bit Score: 63.46 E-value: 1.98e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYIlgkdirsemssIRQNLGVCPQHNVLFDMlTVEEHIWFYARL 1000
Cdd:PLN03232 638 LEIPVGSLVAIVGGTGEGKTSLISAMLGELSHAETSSVV-----------IRGSVAYVPQVSWIFNA-TVRENILFGSDF 705
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 KglSEKHVKA-EMEQMALDVGLPP----SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL 1075
Cdd:PLN03232 706 E--SERYWRAiDVTALQHDLDLLPgrdlTEIGERGVNISGGQKQRVSMARAVYSNSDIYIFDDPLSALDAHVAHQVFDSC 783
|
170 180 190
....*....|....*....|....*....|...
gi 568925948 1076 LKYR-QGRTIILSTHHMDEADILgDRIAIISHG 1107
Cdd:PLN03232 784 MKDElKGKTRVLVTNQLHFLPLM-DRIILVSEG 815
|
|
| PRK11174 |
PRK11174 |
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed |
889-1109 |
2.07e-09 |
|
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed
Pssm-ID: 236870 [Multi-domain] Cd Length: 588 Bit Score: 62.55 E-value: 2.07e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHLRLGVSI--QNLVKVYRDGmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGlFPPTSGTAYILGKDIRS 966
Cdd:PRK11174 338 GEKELASNDPVTIeaEDLEILSPDG-KTLAGPLNFTLPAGQRIALVGPSGAGKTSLLNALLG-FLPYQGSLKINGIELRE 415
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 967 -EMSSIRQNLGVCPQHNVLFDMlTVEEHIwfyarLKG---LSEKHVKAEMEQM-------ALDVGLPpSKLKSKTSQLSG 1035
Cdd:PRK11174 416 lDPESWRKHLSWVGQNPQLPHG-TLRDNV-----LLGnpdASDEQLQQALENAwvseflpLLPQGLD-TPIGDQAAGLSV 488
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1036 GMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILgDRIAIISHGKL 1109
Cdd:PRK11174 489 GQAQRLALARALLQPCQLLLLDEPTASLDAHSEQLVMQALNAASRRQTTLMVTHQLEDLAQW-DQIWVMQDGQI 561
|
|
| PRK10762 |
PRK10762 |
D-ribose transporter ATP binding protein; Provisional |
1913-2124 |
2.27e-09 |
|
D-ribose transporter ATP binding protein; Provisional
Pssm-ID: 236755 [Multi-domain] Cd Length: 501 Bit Score: 62.33 E-value: 2.27e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYrrkrkP---AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGdtpVTRGDA----FLNKNSILSN--- 1982
Cdd:PRK10762 4 LLQLKGIDKAF-----PgvkALSGAALNVYPGRVMALVGENGAGKSTMMKVLTG---IYTRDAgsilYLGKEVTFNGpks 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1983 -----IHEVHQNMGYCPQFDAITELLTGREHVEFFAllrGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLST 2057
Cdd:PRK10762 76 sqeagIGIIHQELNLIPQLTIAENIFLGREFVNRFG---RIDWKKMYAEADKLLARLNLRFSSDKLVGELSIGEQQMVEI 152
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:PRK10762 153 AKVLSFESKVIIMDEPTDALTDTETESLFRVIRELKSQGRGIVYISHRLKEIFEICDDVTVFRDGQF 219
|
|
| ssuB |
PRK11247 |
aliphatic sulfonates transport ATP-binding subunit; Provisional |
890-1109 |
2.43e-09 |
|
aliphatic sulfonates transport ATP-binding subunit; Provisional
Pssm-ID: 183055 [Multi-domain] Cd Length: 257 Bit Score: 60.46 E-value: 2.43e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 890 EEPTHLRLG--VSIQNLVKVYrdGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGtAYILGkdiRSE 967
Cdd:PRK11247 2 MNTARLNQGtpLLLNAVSKRY--GERTVLNQLDLHIPAGQFVAVVGRSGCGKSTLLRLLAGLETPSAG-ELLAG---TAP 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MSSIRQNLGVCPQHNVLFDMLTVEEHIwfyarlkGLSEK-HVKAEMEQmALD-VGLPpSKLKSKTSQLSGGMQRKLSVAL 1045
Cdd:PRK11247 76 LAEAREDTRLMFQDARLLPWKKVIDNV-------GLGLKgQWRDAALQ-ALAaVGLA-DRANEWPAALSGGQKQRVALAR 146
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1046 AFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK11247 147 ALIHRPGLLLLDEPLGALDALTRIEMQDLIesLWQQHGFTVLLVTHDVSEAVAMADRVLLIEEGKI 212
|
|
| PRK13546 |
PRK13546 |
teichoic acids export ABC transporter ATP-binding subunit TagH; |
899-1109 |
2.47e-09 |
|
teichoic acids export ABC transporter ATP-binding subunit TagH;
Pssm-ID: 184131 [Multi-domain] Cd Length: 264 Bit Score: 60.60 E-value: 2.47e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYR------DGMK------------VAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAyil 960
Cdd:PRK13546 5 VNIKNVTKEYRiyrtnkERMKdalipkhknktfFALDDISLKAYEGDVIGLVGINGSGKSTLSNIIGGSLSPTVGKV--- 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 961 gkDIRSEMSSIRQNLGVCPQhnvlfdmLTVEEHIWFYARLKGLSEKHVKAEMEQMaldvgLPPSKLKSKTSQ----LSGG 1036
Cdd:PRK13546 82 --DRNGEVSVIAISAGLSGQ-------LTGIENIEFKMLCMGFKRKEIKAMTPKI-----IEFSELGEFIYQpvkkYSSG 147
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 1037 MQRKLSVALAFVGGSKVVILDEP-TAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK13546 148 MRAKLGFSINITVNPDILVIDEAlSVGDQTFAQKCLDKIYEFKEQNKTIFFVSHNLGQVRQFCTKIAWIEGGKL 221
|
|
| ATM1 |
COG5265 |
ABC-type transport system involved in Fe-S cluster assembly, permease and ATPase components ... |
926-1088 |
2.49e-09 |
|
ABC-type transport system involved in Fe-S cluster assembly, permease and ATPase components [Posttranslational modification, protein turnover, chaperones];
Pssm-ID: 444078 [Multi-domain] Cd Length: 605 Bit Score: 62.53 E-value: 2.49e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGVCPQHNVLF-DmlTVEEHIWfYARLkGL 1003
Cdd:COG5265 384 GKTVAIVGPSGAGKSTLARLLFRFYDVTSGRILIDGQDIRDvTQASLRAAIGIVPQDTVLFnD--TIAYNIA-YGRP-DA 459
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1004 SEkhvkAEMEQMA----LD---VGLPpsK----------LKsktsqLSGG-MQRklsVALA--FVGGSKVVILDEPTAGV 1063
Cdd:COG5265 460 SE----EEVEAAAraaqIHdfiESLP--DgydtrvgergLK-----LSGGeKQR---VAIArtLLKNPPILIFDEATSAL 525
|
170 180 190
....*....|....*....|....*....|
gi 568925948 1064 DPYSRRGIWELLLKYRQGRT--II---LST 1088
Cdd:COG5265 526 DSRTERAIQAALREVARGRTtlVIahrLST 555
|
|
| PRK10261 |
PRK10261 |
glutathione transporter ATP-binding protein; Provisional |
1882-2123 |
2.50e-09 |
|
glutathione transporter ATP-binding protein; Provisional
Pssm-ID: 182342 [Multi-domain] Cd Length: 623 Bit Score: 62.57 E-value: 2.50e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1882 PVKAKLPPLNDED-EDVRRERQRILDGggqNDILEIKELTKIY--------RRKRK-PAVDRICIGIPPGECFGLLGVNG 1951
Cdd:PRK10261 284 PRRFPLISLEHPAkQEPPIEQDTVVDG---EPILQVRNLVTRFplrsgllnRVTREvHAVEKVSFDLWPGETLSLVGESG 360
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1952 AGKSTTFKMLTGDTPVTRGDAFLNKNSI----LSNIHEVHQNMGYCPQ------------FDAITELLtgREHveffALL 2015
Cdd:PRK10261 361 SGKSTTGRALLRLVESQGGEIIFNGQRIdtlsPGKLQALRRDIQFIFQdpyasldprqtvGDSIMEPL--RVH----GLL 434
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2016 RGvpeKEVGKVGEWAIRKLGLV-KYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVK 2094
Cdd:PRK10261 435 PG---KAAAARVAWLLERVGLLpEHAWRYPHEFSGGQRQRICIARALALNPKVIIADEAVSALDVSIRGQIINLLLDLQR 511
|
250 260 270
....*....|....*....|....*....|
gi 568925948 2095 E-GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK10261 512 DfGIAYLFISHDMAVVERISHRVAVMYLGQ 541
|
|
| ABC_ABC_ChvD |
TIGR03719 |
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of ... |
1890-2078 |
3.41e-09 |
|
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of the ABC transporter ATP-binding cassette, but are found outside the common ABC transporter operon structure that features integral membrane permease proteins and substrate-binding proteins encoded next to the ATP-binding cassette (ABC domain) protein. The member protein ChvD from Agrobacterium tumefaciens was identified as both a candidate to interact with VirB8, based on yeast two-hybrid analysis, and as an apparent regulator of VirG. The general function of this protein family is unknown.
Pssm-ID: 274744 [Multi-domain] Cd Length: 552 Bit Score: 61.87 E-value: 3.41e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1890 LNDEDEDVRRERQRILDGGGQ---NDILEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DT 1965
Cdd:TIGR03719 296 LLSQEFQKRNETAEIYIPPGPrlgDKVIEAENLTKAF--GDKLLIDDLSFKLPPGGIVGVIGPNGAGKSTLFRMITGqEQ 373
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1966 P----VTRGDAflnknsilsniheVHqnMGYCPQFdaiTELLTGREHVeFFALLRGVPEKEVGKVgEWAIRK-LGLVKYG 2040
Cdd:TIGR03719 374 PdsgtIEIGET-------------VK--LAYVDQS---RDALDPNKTV-WEEISGGLDIIKLGKR-EIPSRAyVGRFNFK 433
|
170 180 190 200
....*....|....*....|....*....|....*....|..
gi 568925948 2041 ----EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:TIGR03719 434 gsdqQKKVGQLSGGERNRVHLAKTLKSGGNVLLLDEPTNDLD 475
|
|
| PRK10619 |
PRK10619 |
histidine ABC transporter ATP-binding protein HisP; |
1910-2107 |
3.62e-09 |
|
histidine ABC transporter ATP-binding protein HisP;
Pssm-ID: 182592 [Multi-domain] Cd Length: 257 Bit Score: 59.98 E-value: 3.62e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNsilsNIHEVHQN 1989
Cdd:PRK10619 2 SENKLNVIDLHKRYGEHE--VLKGVSLQANAGDVISIIGSSGSGKSTFLRCINFLEKPSEGSIVVNGQ----TINLVRDK 75
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQFDAIT-ELLTGR-----EHV---EFFALLRGVPEKEVGKVG-------EWAIRKLGLVKYGE----KYASNYSG 2049
Cdd:PRK10619 76 DGQLKVADKNQlRLLRTRltmvfQHFnlwSHMTVLENVMEAPIQVLGlskqearERAVKYLAKVGIDEraqgKYPVHLSG 155
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2050 GNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSME 2107
Cdd:PRK10619 156 GQQQRVSIARALAMEPEVLLFDEPTSALDPELVGEVLRIMQQLAEEGKTMVVVTHEMG 213
|
|
| CeuD |
COG4604 |
ABC-type enterochelin transport system, ATPase component [Inorganic ion transport and ... |
1915-2104 |
4.58e-09 |
|
ABC-type enterochelin transport system, ATPase component [Inorganic ion transport and metabolism];
Pssm-ID: 443654 [Multi-domain] Cd Length: 252 Bit Score: 59.33 E-value: 4.58e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1915 EIKELTKIYRRKrkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYC 1993
Cdd:COG4604 3 EIKNVSKRYGGK--VVLDDVSLTIPKGGITALIGPNGAGKSTLLSMISRLLPPDSGEVLVDGLDVATtPSRELAKRLAIL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEF--FALLRGVPEKE-VGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:COG4604 81 RQENHINSRLTVRELVAFgrFPYSKGRLTAEdREIIDE-AIAYLDLEDLADRYLDELSGGQRQRAFIAMVLAQDTDYVLL 159
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 2071 DEPTTGMDPKA--------RRflwncalsIVKE-GRSVVLTSH 2104
Cdd:COG4604 160 DEPLNNLDMKHsvqmmkllRR--------LADElGKTVVIVLH 194
|
|
| GguA |
NF040905 |
sugar ABC transporter ATP-binding protein; |
911-1108 |
5.01e-09 |
|
sugar ABC transporter ATP-binding protein;
Pssm-ID: 468840 [Multi-domain] Cd Length: 500 Bit Score: 61.34 E-value: 5.01e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYIL--G-----KDIR-SEMSS---IRQNLGVCP 979
Cdd:NF040905 13 GVK-ALDDVNLSVREGEIHALCGENGAGKSTLMKVLSGVYPHGSYEGEILfdGevcrfKDIRdSEALGiviIHQELALIP 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 980 QhnvlfdmLTVEEHIWF---YARlKGL---SEKHVKAEmEQMAlDVGL--PPSklkSKTSQLSGGMQRKLSVALAFVGGS 1051
Cdd:NF040905 92 Y-------LSIAENIFLgneRAK-RGVidwNETNRRAR-ELLA-KVGLdeSPD---TLVTDIGVGKQQLVEIAKALSKDV 158
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 1052 KVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMDEADILGDRIAIISHGK 1108
Cdd:NF040905 159 KLLILDEPTAALNEEDSAALLDLLLELKaQGITSIIISHKLNEIRRVADSITVLRDGR 216
|
|
| PRK10261 |
PRK10261 |
glutathione transporter ATP-binding protein; Provisional |
899-1114 |
5.16e-09 |
|
glutathione transporter ATP-binding protein; Provisional
Pssm-ID: 182342 [Multi-domain] Cd Length: 623 Bit Score: 61.41 E-value: 5.16e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNL-VKVYRDGMKV-AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYI--------------LGK 962
Cdd:PRK10261 13 LAVENLnIAFMQEQQKIaAVRNLSFSLQRGETLAIVGESGSGKSVTALALMRLLEQAGGLVQCdkmllrrrsrqvieLSE 92
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 963 DIRSEMSSIR-QNLGVCPQHNV--LFDMLTVEEHIWFYARL-KGLSEKHVKAEMEQMALDVGLPPSK--LKSKTSQLSGG 1036
Cdd:PRK10261 93 QSAAQMRHVRgADMAMIFQEPMtsLNPVFTVGEQIAESIRLhQGASREEAMVEAKRMLDQVRIPEAQtiLSRYPHQLSGG 172
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1037 MQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHMDEADILGDRIAIISHGKLCCVGS 1114
Cdd:PRK10261 173 MRQRVMIAMALSCRPAVLIADEPTTALDVTIQAQILQLIkvLQKEMSMGVIFITHDMGVVAEIADRVLVMYQGEAVETGS 252
|
|
| cbiO |
PRK13646 |
energy-coupling factor transporter ATPase; |
1917-2122 |
5.83e-09 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184205 [Multi-domain] Cd Length: 286 Bit Score: 59.79 E-value: 5.83e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1917 KELTKIYRRK---RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSN-----IHEVHQ 1988
Cdd:PRK13646 6 DNVSYTYQKGtpyEHQAIHDVNTEFEQGKYYAIVGQTGSGKSTLIQNINALLKPTTGTVTVDDITITHKtkdkyIRPVRK 85
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQF--DAITELLTGREhVEFFALLRGVPEKEVGKVGEWAIRKLGLVK-YGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:PRK13646 86 RIGMVFQFpeSQLFEDTVERE-IIFGPKNFKMNLDEVKNYAHRLLMDLGFSRdVMSQSPFQMSGGQMRKIAIVSILAMNP 164
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2066 PVVFLDEPTTGMDPKARRFLWNCALSI-VKEGRSVVLTSHSMEECEALCTRMAIMVNG 2122
Cdd:PRK13646 165 DIIVLDEPTAGLDPQSKRQVMRLLKSLqTDENKTIILVSHDMNEVARYADEVIVMKEG 222
|
|
| PRK10851 |
PRK10851 |
sulfate/thiosulfate ABC transporter ATP-binding protein CysA; |
1914-2131 |
6.18e-09 |
|
sulfate/thiosulfate ABC transporter ATP-binding protein CysA;
Pssm-ID: 182778 [Multi-domain] Cd Length: 353 Bit Score: 60.10 E-value: 6.18e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIHEVHQNMGYC 1993
Cdd:PRK10851 3 IEIANIKKSFGRTQ--VLNDISLDIPSGQMVALLGPSGSGKTTLLRIIAGLEHQTSGHIRFHGTDV-SRLHARDRKVGFV 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQFDAITELLTGREHVEFFalLRGVPEKEvgKVGEWAIRK-----LGLVKYG---EKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:PRK10851 80 FQHYALFRHMTVFDNIAFG--LTVLPRRE--RPNAAAIKAkvtqlLEMVQLAhlaDRYPAQLSGGQKQRVALARALAVEP 155
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2066 PVVFLDEPTTGMDPKARRFL--WNCALSIVKEGRSVVLTsHSMEECEALCTRMAIMVNGRFRCLGSVQ 2131
Cdd:PRK10851 156 QILLLDEPFGALDAQVRKELrrWLRQLHEELKFTSVFVT-HDQEEAMEVADRVVVMSQGNIEQAGTPD 222
|
|
| cbiO |
PRK13645 |
energy-coupling factor transporter ATPase; |
1946-2129 |
7.51e-09 |
|
energy-coupling factor transporter ATPase;
Pssm-ID: 184204 [Multi-domain] Cd Length: 289 Bit Score: 59.25 E-value: 7.51e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1946 LLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQ------NMGYC---PQFDAITEllTGREHVEFFALLR 2016
Cdd:PRK13645 42 VIGTTGSGKSTMIQLTNGLIISETGQTIVGDYAIPANLKKIKEvkrlrkEIGLVfqfPEYQLFQE--TIEKDIAFGPVNL 119
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2017 GVPEKEV-GKVGEWairkLGLVKYGEKYAS----NYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALS 2091
Cdd:PRK13645 120 GENKQEAyKKVPEL----LKLVQLPEDYVKrspfELSGGQKRRVALAGIIAMDGNTLVLDEPTGGLDPKGEEDFINLFER 195
|
170 180 190
....*....|....*....|....*....|....*....
gi 568925948 2092 IVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGS 2129
Cdd:PRK13645 196 LNKEyKKRIIMVTHNMDQVLRIADEVIVMHEGKVISIGS 234
|
|
| araG |
PRK11288 |
L-arabinose ABC transporter ATP-binding protein AraG; |
1930-2124 |
7.68e-09 |
|
L-arabinose ABC transporter ATP-binding protein AraG;
Pssm-ID: 183077 [Multi-domain] Cd Length: 501 Bit Score: 60.70 E-value: 7.68e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN---------KNSILSNIHEVHQNMGYCPQFDAIT 2000
Cdd:PRK11288 19 ALDDISFDCRAGQVHALMGENGAGKSTLLKILSGNYQPDAGSILIDgqemrfastTAALAAGVAIIYQELHLVPEMTVAE 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 ELLTGREHVEFFALLRGVPEKEVGKvgewAIRKLGL-------VKYgekyasnYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:PRK11288 99 NLYLGQLPHKGGIVNRRLLNYEARE----QLEHLGVdidpdtpLKY-------LSIGQRQMVEIAKALARNARVIAFDEP 167
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2074 TTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:PRK11288 168 TSSLSAREIEQLFRVIRELRAEGRVILYVSHRMEEIFALCDAITVFKDGRY 218
|
|
| cbiO |
PRK13631 |
cobalt transporter ATP-binding subunit; Provisional |
1911-2107 |
9.06e-09 |
|
cobalt transporter ATP-binding subunit; Provisional
Pssm-ID: 237451 [Multi-domain] Cd Length: 320 Bit Score: 59.48 E-value: 9.06e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIYRRKRK---PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDA----FLNKNSILSNI 1983
Cdd:PRK13631 19 DIILRVKNLYCVFDEKQEnelVALNNISYTFEKNKIYFIIGNSGSGKSTLVTHFNGLIKSKYGTIqvgdIYIGDKKNNHE 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1984 HEVHQNMGYCPQFDAITELL--------------TGREHVEFFALLRGVPEKEVGKVGEWAIRKLGL-VKYGEKYASNYS 2048
Cdd:PRK13631 99 LITNPYSKKIKNFKELRRRVsmvfqfpeyqlfkdTIEKDIMFGPVALGVKKSEAKKLAKFYLNKMGLdDSYLERSPFGLS 178
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2049 GGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSME 2107
Cdd:PRK13631 179 GGQKRRVAIAGILAIQPEILIFDEPTAGLDPKGEHEMMQLILDAKANNKTVFVITHTME 237
|
|
| ABCG_PDR_domain1 |
cd03233 |
First domain of the pleiotropic drug resistance-like subfamily G of ATP-binding cassette ... |
1940-2078 |
9.22e-09 |
|
First domain of the pleiotropic drug resistance-like subfamily G of ATP-binding cassette transporters; The pleiotropic drug resistance (PDR) is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. This PDR subfamily represents domain I of its (ABC-IM)2 organization. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds including sugars, ions, peptides, and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213200 [Multi-domain] Cd Length: 202 Bit Score: 57.66 E-value: 9.22e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1940 PGECFGLLGVNGAGKSTTFKMLTGDTPVT---RGDAFLNKNSILSNIHEVHQNMGYCPQFDAITELLTGREHVEFFALLR 2016
Cdd:cd03233 32 PGEMVLVLGRPGSGCSTLLKALANRTEGNvsvEGDIHYNGIPYKEFAEKYPGEIIYVSEEDVHFPTLTVRETLDFALRCK 111
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2017 GvpekevgkvgewairklglvkygEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:cd03233 112 G-----------------------NEFVRGISGGERKRVSIAEALVSRASVLCWDNSTRGLD 150
|
|
| phnK |
PRK11701 |
phosphonate C-P lyase system protein PhnK; Provisional |
1913-2123 |
9.56e-09 |
|
phosphonate C-P lyase system protein PhnK; Provisional
Pssm-ID: 183280 [Multi-domain] Cd Length: 258 Bit Score: 58.78 E-value: 9.56e-09
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIY-RRKrkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDA-FLNKNSILSNIHE----- 1985
Cdd:PRK11701 6 LLSVRGLTKLYgPRK---GCRDVSFDLYPGEVLGIVGESGSGKTTLLNALSARLAPDAGEVhYRMRDGQLRDLYAlseae 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 -----------VHQN--MGYCPQFDA---ITElltgRehveffalLRGVPEKEVGKVGEWAIRKLGLVKYGEK----YAS 2045
Cdd:PRK11701 83 rrrllrtewgfVHQHprDGLRMQVSAggnIGE----R--------LMAVGARHYGDIRATAGDWLERVEIDAAriddLPT 150
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2046 NYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD--PKARrfLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNG 2122
Cdd:PRK11701 151 TFSGGMQQRLQIARNLVTHPRLVFMDEPTGGLDvsVQAR--LLDLLRGLVRElGLAVVIVTHDLAVARLLAHRLLVMKQG 228
|
.
gi 568925948 2123 R 2123
Cdd:PRK11701 229 R 229
|
|
| ABCG_PDR_domain1 |
cd03233 |
First domain of the pleiotropic drug resistance-like subfamily G of ATP-binding cassette ... |
921-1064 |
1.01e-08 |
|
First domain of the pleiotropic drug resistance-like subfamily G of ATP-binding cassette transporters; The pleiotropic drug resistance (PDR) is a well-described phenomenon occurring in fungi and shares several similarities with processes in bacteria and higher eukaryotes. This PDR subfamily represents domain I of its (ABC-IM)2 organization. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds including sugars, ions, peptides, and more complex organic molecules. The nucleotide-binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213200 [Multi-domain] Cd Length: 202 Bit Score: 57.66 E-value: 1.01e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFY--EGQITSFLGHNGAGKTTTMSILTGLFPPT---SGTAYILGKDIRSEMSSIRQNLGVCPQHNVLFDMLTVEEHIW 995
Cdd:cd03233 26 FSGVvkPGEMVLVLGRPGSGCSTLLKALANRTEGNvsvEGDIHYNGIPYKEFAEKYPGEIIYVSEEDVHFPTLTVRETLD 105
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 996 FYARLKGlsekhvkaemEQMaldvglppsklkskTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVD 1064
Cdd:cd03233 106 FALRCKG----------NEF--------------VRGISGGERKRVSIAEALVSRASVLCWDNSTRGLD 150
|
|
| tagH |
PRK13545 |
teichoic acids export protein ATP-binding subunit; Provisional |
1930-2139 |
1.03e-08 |
|
teichoic acids export protein ATP-binding subunit; Provisional
Pssm-ID: 184130 [Multi-domain] Cd Length: 549 Bit Score: 60.29 E-value: 1.03e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEvhqnmgycpqfdAITELLTGREHV 2009
Cdd:PRK13545 39 ALNNISFEVPEGEIVGIIGLNGSGKSTLSNLIAGVTMPNKGTVDIKGSAALIAISS------------GLNGQLTGIENI 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2010 EFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDpkaRRFLWNC- 2088
Cdd:PRK13545 107 ELKGLMMGLTKEKIKEIIPEIIEFADIGKFIYQPVKTYSSGMKSRLGFAISVHINPDILVIDEALSVGD---QTFTKKCl 183
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2089 -ALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFGD 2139
Cdd:PRK13545 184 dKMNEFKEqGKTIFFISHSLSQVKSFCTKALWLHYGQVKEYGDIKEVVDHYDE 236
|
|
| ABC_RNaseL_inhibitor_domain1 |
cd03236 |
The ATP-binding cassette domain 1 of RNase L inhibitor; The ABC ATPase, RNase L inhibitor (RLI) ... |
1940-2104 |
1.05e-08 |
|
The ATP-binding cassette domain 1 of RNase L inhibitor; The ABC ATPase, RNase L inhibitor (RLI), is a key enzyme in ribosomal biogenesis, formation of translation preinitiation complexes, and assembly of HIV capsids. RLI s are not transport proteins and thus cluster with a group of soluble proteins that lack the transmembrane components commonly found in other members of the family. Structurally, RLIs have an N-terminal Fe-S domain and two nucleotide binding domains which are arranged to form two composite active sites in their interface cleft. RLI is one of the most conserved enzymes between archaea and eukaryotes with a sequence identity more than 48%. The high degree of evolutionary conservation suggests that RLI performs a central role in archaeal and eukaryotic physiology.
Pssm-ID: 213203 [Multi-domain] Cd Length: 255 Bit Score: 58.53 E-value: 1.05e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1940 PGECFGLLGVNGAGKSTTFKMLTGdtpvtrgdaflnknsilsnihEVHQNMG-YC--PQFDAITELLTGREHVEFFALLR 2016
Cdd:cd03236 25 EGQVLGLVGPNGIGKSTALKILAG---------------------KLKPNLGkFDdpPDWDEILDEFRGSELQNYFTKLL 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2017 G--------------VPEKEVGKVGE------------WAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFL 2070
Cdd:cd03236 84 EgdvkvivkpqyvdlIPKAVKGKVGEllkkkdergkldELVDQLELRHVLDRNIDQLSGGELQRVAIAAALARDADFYFF 163
|
170 180 190
....*....|....*....|....*....|....
gi 568925948 2071 DEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:cd03236 164 DEPSSYLDIKQRLNAARLIRELAEDDNYVLVVEH 197
|
|
| xylG |
TIGR02633 |
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose ... |
1911-2125 |
1.31e-08 |
|
D-xylose ABC transporter, ATP-binding protein; Several bacterial species have enzymes xylose isomerase and xylulokinase enzymes for xylose utilization. Members of this protein family are the ATP-binding cassette (ABC) subunit of the known or predicted high-affinity xylose ABC transporter for xylose import. These genes, which closely resemble other sugar transport ABC transporter genes, typically are encoded near xylose utilization enzymes and regulatory proteins. Note that this form of the transporter contains two copies of the ABC transporter domain (pfam00005). [Transport and binding proteins, Carbohydrates, organic alcohols, and acids]
Pssm-ID: 131681 [Multi-domain] Cd Length: 500 Bit Score: 59.84 E-value: 1.31e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1911 NDILEIKELTKIY----RRKRkpaVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP-VTRGDAFLNKNSI------ 1979
Cdd:TIGR02633 255 DVILEARNLTCWDvinpHRKR---VDDVSFSLRRGEILGVAGLVGAGRTELVQALFGAYPgKFEGNVFINGKPVdirnpa 331
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1980 ------LSNIHEVHQNMGYCPQFdAITELLTGREHVEFFALLRGVPEKEVGKVGEwAIRKLGLvkygeKYASNY------ 2047
Cdd:TIGR02633 332 qairagIAMVPEDRKRHGIVPIL-GVGKNITLSVLKSFCFKMRIDAAAELQIIGS-AIQRLKV-----KTASPFlpigrl 404
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2048 SGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFR 2125
Cdd:TIGR02633 405 SGGNQQKAVLAKMLLTNPRVLILDEPTRGVDVGAKYEIYKLINQLAQEGVAIIVVSSELAEVLGLSDRVLVIGEGKLK 482
|
|
| PRK10982 |
PRK10982 |
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional |
1913-2123 |
1.31e-08 |
|
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional
Pssm-ID: 182880 [Multi-domain] Cd Length: 491 Bit Score: 59.74 E-value: 1.31e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKiyrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL-SNIHEVHQNmg 1991
Cdd:PRK10982 250 ILEVRNLTS----LRQPSIRDVSFDLHKGEILGIAGLVGAKRTDIVETLFGIREKSAGTITLHGKKINnHNANEAINH-- 323
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 ycpQFDAITEL-----LTGREHVEFFALLRGVpEKEVGKVG-----------EWAIRKLGLVKYGEKYA-SNYSGGNKRK 2054
Cdd:PRK10982 324 ---GFALVTEErrstgIYAYLDIGFNSLISNI-RNYKNKVGlldnsrmksdtQWVIDSMRVKTPGHRTQiGSLSGGNQQK 399
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2055 LSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK10982 400 VIIGRWLLTQPEILMLDEPTRGIDVGAKFEIYQLIAELAKKDKGIIIISSEMPELLGITDRILVMSNGL 468
|
|
| MRP_assoc_pro |
TIGR00957 |
multi drug resistance-associated protein (MRP); This model describes multi drug ... |
899-1114 |
1.56e-08 |
|
multi drug resistance-associated protein (MRP); This model describes multi drug resistance-associated protein (MRP) in eukaryotes. The multidrug resistance-associated protein is an integral membrane protein that causes multidrug resistance when overexpressed in mammalian cells. It belongs to ABC transporter superfamily. The protein topology and function was experimentally demonstrated by epitope tagging and immunofluorescence. Insertion of tags in the critical regions associated with drug efflux, abrogated its function. The C-terminal domain seem to highly conserved. [Transport and binding proteins, Other]
Pssm-ID: 188098 [Multi-domain] Cd Length: 1522 Bit Score: 60.34 E-value: 1.56e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNLGVc 978
Cdd:TIGR00957 637 ITVHNATFTWARDLPPTLNGITFSIPEGALVAVVGQVGCGKSSLLSALLAEMDKVEGHVHMKGSVAYVPQQAWIQNDSL- 715
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 979 pQHNVLFDmltveehiwfyarlKGLSEKHVKAEMEQMAL--DVGLPPS----KLKSKTSQLSGGMQRKLSVALAFVGGSK 1052
Cdd:TIGR00957 716 -RENILFG--------------KALNEKYYQQVLEACALlpDLEILPSgdrtEIGEKGVNLSGGQKQRVSLARAVYSNAD 780
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1053 VVILDEPTAGVDPYSRRGIWELLLKYR---QGRTIILSTHHMDEADILgDRIAIISHGKLCCVGS 1114
Cdd:TIGR00957 781 IYLFDDPLSAVDAHVGKHIFEHVIGPEgvlKNKTRILVTHGISYLPQV-DVIIVMSGGKISEMGS 844
|
|
| PRK14271 |
PRK14271 |
phosphate ABC transporter ATP-binding protein; Provisional |
1928-2133 |
1.66e-08 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172759 [Multi-domain] Cd Length: 276 Bit Score: 58.18 E-value: 1.66e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPVT----RGDAFLNKNSILS--NIHEVHQNMGYCPQ----- 1995
Cdd:PRK14271 34 KTVLDQVSMGFPARAVTSLMGPTGSGKTTFLRTLNRmNDKVSgyrySGDVLLGGRSIFNyrDVLEFRRRVGMLFQrpnpf 113
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1996 -FDAITELLTG-REHveffallRGVPEKEVGKVGEWAIRKLGLVKYGEKYASN----YSGGNKRKLSTAMALIGGPPVVF 2069
Cdd:PRK14271 114 pMSIMDNVLAGvRAH-------KLVPRKEFRGVAQARLTEVGLWDAVKDRLSDspfrLSGGQQQLLCLARTLAVNPEVLL 186
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2070 LDEPTTGMDP----KARRFLWNCALSIvkegrSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK14271 187 LDEPTSALDPttteKIEEFIRSLADRL-----TVIIVTHNLAQAARISDRAALFFDGRLVEEGPTEQL 249
|
|
| PRK13540 |
PRK13540 |
cytochrome c biogenesis protein CcmA; Provisional |
926-1090 |
1.71e-08 |
|
cytochrome c biogenesis protein CcmA; Provisional
Pssm-ID: 184127 [Multi-domain] Cd Length: 200 Bit Score: 56.88 E-value: 1.71e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIRQNL-------GVCP----QHNVLFDMLTVEEHI 994
Cdd:PRK13540 27 GGLLHLKGSNGAGKTTLLKLIAGLLNPEKGEILFERQSIKKDLCTYQKQLcfvghrsGINPyltlRENCLYDIHFSPGAV 106
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 995 wfyarlkGLSEKHVKAEMEQMaLDVglpPSKLksktsqLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWEL 1074
Cdd:PRK13540 107 -------GITELCRLFSLEHL-IDY---PCGL------LSSGQKRQVALLRLWMSKAKLWLLDEPLVALDELSLLTIITK 169
|
170
....*....|....*..
gi 568925948 1075 LLKYR-QGRTIILSTHH 1090
Cdd:PRK13540 170 IQEHRaKGGAVLLTSHQ 186
|
|
| PRK11160 |
PRK11160 |
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed |
1914-2133 |
1.76e-08 |
|
cysteine/glutathione ABC transporter membrane/ATP-binding component; Reviewed
Pssm-ID: 236865 [Multi-domain] Cd Length: 574 Bit Score: 59.84 E-value: 1.76e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNIHE--VHQNMG 1991
Cdd:PRK11160 339 LTLNNVSFTYPDQPQPVLKGLSLQIKAGEKVALLGRTGCGKSTLLQLLTRAWDPQQGEILLNGQPI-ADYSEaaLRQAIS 417
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQfdaitelltgREHVeFFALLR-----GVPEKEVGKVGEwAIRKLGLvkygEKYASNY--------------SGGNK 2052
Cdd:PRK11160 418 VVSQ----------RVHL-FSATLRdnlllAAPNASDEALIE-VLQQVGL----EKLLEDDkglnawlgeggrqlSGGEQ 481
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2053 RKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLwncaLSIVKE---GRSVVLTSH---SMEECEALCtrmaIMVNGRFRC 2126
Cdd:PRK11160 482 RRLGIARALLHDAPLLLLDEPTEGLDAETERQI----LELLAEhaqNKTVLMITHrltGLEQFDRIC----VMDNGQIIE 553
|
....*..
gi 568925948 2127 LGSVQHL 2133
Cdd:PRK11160 554 QGTHQEL 560
|
|
| nikD |
PRK10418 |
nickel transporter ATP-binding protein NikD; Provisional |
1928-2136 |
2.03e-08 |
|
nickel transporter ATP-binding protein NikD; Provisional
Pssm-ID: 236688 [Multi-domain] Cd Length: 254 Bit Score: 57.40 E-value: 2.03e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP----VTRGDAFLNKNSILSN------IHEVHQNmgycPQfD 1997
Cdd:PRK10418 16 QPLVHGVSLTLQRGRVLALVGGSGSGKSLTCAAALGILPagvrQTAGRVLLDGKPVAPCalrgrkIATIMQN----PR-S 90
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1998 AITELLTGREHVEFFALLRGVPEKEVGKVGewAIRKLGLVKYG---EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPT 2074
Cdd:PRK10418 91 AFNPLHTMHTHARETCLALGKPADDATLTA--ALEAVGLENAArvlKLYPFEMSGGMLQRMMIALALLCEAPFIIADEPT 168
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2075 TGMDPKAR-RFLwNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNR 2136
Cdd:PRK10418 169 TDLDVVAQaRIL-DLLESIVQKrALGMLLVTHDMGVVARLADDVAVMSHGRIVEQGDVETLFNA 231
|
|
| oppD |
PRK09473 |
oligopeptide transporter ATP-binding component; Provisional |
910-1089 |
2.11e-08 |
|
oligopeptide transporter ATP-binding component; Provisional
Pssm-ID: 181888 [Multi-domain] Cd Length: 330 Bit Score: 58.58 E-value: 2.11e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 910 DGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPP---TSGTAYILGKDI-----------RSE-------- 967
Cdd:PRK09473 26 DGDVTAVNDLNFSLRAGETLGIVGESGSGKSQTAFALMGLLAAngrIGGSATFNGREIlnlpekelnklRAEqismifqd 105
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 -MSSIRQNLGVCPQ-HNVLfdMLtveeHiwfyarlKGLSEKHVKAEMEQMALDVGLPPSKLKSKT--SQLSGGMQRKLSV 1043
Cdd:PRK09473 106 pMTSLNPYMRVGEQlMEVL--ML----H-------KGMSKAEAFEESVRMLDAVKMPEARKRMKMypHEFSGGMRQRVMI 172
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTH 1089
Cdd:PRK09473 173 AMALLCRPKLLIADEPTTALDVTVQAQIMTLLneLKREFNTAIIMITH 220
|
|
| ABCC_MRP_domain1 |
cd03250 |
ATP-binding cassette domain 1 of multidrug resistance-associated protein, subfamily C; This ... |
921-1108 |
2.18e-08 |
|
ATP-binding cassette domain 1 of multidrug resistance-associated protein, subfamily C; This subfamily is also known as MRP (multidrug resistance-associated protein). Some of the MRP members have five additional transmembrane segments in their N-terminus, but the function of these additional membrane-spanning domains is not clear. The MRP was found in the multidrug-resisting lung cancer cell in which p-glycoprotein was not overexpressed. MRP exports glutathione by drug stimulation, as well as, certain substrates in conjugated forms with anions, such as glutathione, glucuronate, and sulfate.
Pssm-ID: 213217 [Multi-domain] Cd Length: 204 Bit Score: 56.71 E-value: 2.18e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKdirsemssirqnLGVCPQHNVLFDMlTVEEHIWFYARL 1000
Cdd:cd03250 26 LEVPKGELVAIVGPVGSGKSSLLSALLGELEKLSGSVSVPGS------------IAYVSQEPWIQNG-TIRENILFGKPF 92
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 -KGLSEKHVKA-----EMEQMAL----DVGlppsklkSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRG 1070
Cdd:cd03250 93 dEERYEKVIKAcalepDLEILPDgdltEIG-------EKGINLSGGQKQRISLARAVYSDADIYLLDDPLSAVDAHVGRH 165
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 1071 IWELLL--KYRQGRTIILSTHHMD---EAdilgDRIAIISHGK 1108
Cdd:cd03250 166 IFENCIlgLLLNNKTRILVTHQLQllpHA----DQIVVLDNGR 204
|
|
| NupO |
COG3845 |
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and ... |
1913-2124 |
2.36e-08 |
|
ABC-type guanosine uptake system NupNOPQ, ATPase component NupO [Nucleotide transport and metabolism];
Pssm-ID: 443055 [Multi-domain] Cd Length: 504 Bit Score: 59.27 E-value: 2.36e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTkIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI------------L 1980
Cdd:COG3845 257 VLEVENLS-VRDDRGVPALKDVSLEVRAGEILGIAGVAGNGQSELAEALAGLRPPASGSIRLDGEDItglsprerrrlgV 335
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1981 SNIHEVHQNMGYCPQFDaITE--LLTGREHVEF--FALLRGvpekevGKVGEWAIRKLglvkygEKY----------ASN 2046
Cdd:COG3845 336 AYIPEDRLGRGLVPDMS-VAEnlILGRYRRPPFsrGGFLDR------KAIRAFAEELI------EEFdvrtpgpdtpARS 402
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2047 YSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRF 2124
Cdd:COG3845 403 LSGGNQQKVILARELSRDPKLLIAAQPTRGLDVGAIEFIHQRLLELRDAGAAVLLISEDLDEILALSDRIAVMYEGRI 480
|
|
| PLN03211 |
PLN03211 |
ABC transporter G-25; Provisional |
926-1090 |
2.52e-08 |
|
ABC transporter G-25; Provisional
Pssm-ID: 215634 [Multi-domain] Cd Length: 659 Bit Score: 59.12 E-value: 2.52e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDiRSEMSSIRQNLGVCPQHNVLFDMLTVEEHIWFYARL---KG 1002
Cdd:PLN03211 94 GEILAVLGPSGSGKSTLLNALAGRIQGNNFTGTILANN-RKPTKQILKRTGFVTQDDILYPHLTVRETLVFCSLLrlpKS 172
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1003 LSEKHVKAEMEQMALDVGLPPSKL----KSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSR-RGIWELLLK 1077
Cdd:PLN03211 173 LTKQEKILVAESVISELGLTKCENtiigNSFIRGISGGERKRVSIAHEMLINPSLLILDEPTSGLDATAAyRLVLTLGSL 252
|
170
....*....|...
gi 568925948 1078 YRQGRTIILSTHH 1090
Cdd:PLN03211 253 AQKGKTIVTSMHQ 265
|
|
| PLN03140 |
PLN03140 |
ABC transporter G family member; Provisional |
923-1064 |
2.54e-08 |
|
ABC transporter G family member; Provisional
Pssm-ID: 215599 [Multi-domain] Cd Length: 1470 Bit Score: 59.86 E-value: 2.54e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 923 FYEGQITSFLGHNGAGKTTTMSILTGlfPPTSGtaYILGkDIRSEMSSIRQNL-----GVCPQHNVLFDMLTVEEHIWFY 997
Cdd:PLN03140 903 FRPGVLTALMGVSGAGKTTLMDVLAG--RKTGG--YIEG-DIRISGFPKKQETfarisGYCEQNDIHSPQVTVRESLIYS 977
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 998 ARLK-----GLSEK--HVKAEMEQMALD------VGLPpsklksKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVD 1064
Cdd:PLN03140 978 AFLRlpkevSKEEKmmFVDEVMELVELDnlkdaiVGLP------GVTGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLD 1051
|
|
| YnjD |
COG4136 |
ABC-type uncharacterized transport system YnjBCD, ATPase component [General function ... |
1914-2111 |
3.06e-08 |
|
ABC-type uncharacterized transport system YnjBCD, ATPase component [General function prediction only];
Pssm-ID: 443311 [Multi-domain] Cd Length: 211 Bit Score: 56.34 E-value: 3.06e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTkIYRRKRkPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP---VTRGDAFLNKNSILS-NIHEvhQN 1989
Cdd:COG4136 2 LSLENLT-ITLGGR-PLLAPLSLTVAPGEILTLMGPSGSGKSTLLAAIAGTLSpafSASGEVLLNGRRLTAlPAEQ--RR 77
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1990 MGYCPQfdaiTELLtgrehveF----------FALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAM 2059
Cdd:COG4136 78 IGILFQ----DDLL-------FphlsvgenlaFALPPTIGRAQRRARVEQALEEAGLAGFADRDPATLSGGQRARVALLR 146
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2060 ALIGGPPVVFLDEPTTGMDP----KARRFLWNcalSIVKEGRSVVLTSHSMEECEA 2111
Cdd:COG4136 147 ALLAEPRALLLDEPFSKLDAalraQFREFVFE---QIRQRGIPALLVTHDEEDAPA 199
|
|
| bacteriocin_ABC |
TIGR01193 |
ABC-type bacteriocin transporter; This model describes ABC-type bacteriocin transporter. The ... |
1929-2078 |
3.15e-08 |
|
ABC-type bacteriocin transporter; This model describes ABC-type bacteriocin transporter. The amino terminal domain (pfam03412) processes the N-terminal leader peptide from the bacteriocin while C-terminal domains resemble ABC transporter membrane protein and ATP-binding cassette domain. In general, bacteriocins are agents which are responsible for killing or inhibiting the closely related species or even different strains of the same species. Bacteriocins are usually encoded by bacterial plasmids. Bacteriocins are named after the species and hence in literature one encounters various names e.g., leucocin from Leuconostic geldium; pedicocin from Pedicoccus acidilactici; sakacin from Lactobacillus sake etc. [Protein fate, Protein and peptide secretion and trafficking, Protein fate, Protein modification and repair, Transport and binding proteins, Other]
Pssm-ID: 130261 [Multi-domain] Cd Length: 708 Bit Score: 58.98 E-value: 3.15e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1929 PAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlSNI--HEVHQNMGYCPQ----FD-AITE 2001
Cdd:TIGR01193 488 NILSDISLTIKMNSKTTIVGMSGSGKSTLAKLLVGFFQARSGEILLNGFSL-KDIdrHTLRQFINYLPQepyiFSgSILE 566
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2002 -LLTGREhveffallRGVPEKEVGKVGEWA-IRK------LGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEP 2073
Cdd:TIGR01193 567 nLLLGAK--------ENVSQDEIWAACEIAeIKDdienmpLGYQTELSEEGSSISGGQKQRIALARALLTDSKVLILDES 638
|
....*
gi 568925948 2074 TTGMD 2078
Cdd:TIGR01193 639 TSNLD 643
|
|
| PRK14239 |
PRK14239 |
phosphate transporter ATP-binding protein; Provisional |
1913-2122 |
3.15e-08 |
|
phosphate transporter ATP-binding protein; Provisional
Pssm-ID: 184585 [Multi-domain] Cd Length: 252 Bit Score: 57.09 E-value: 3.15e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLT--GDtpvtrgdafLNKN-----SILSNIHE 1985
Cdd:PRK14239 5 ILQVSDLSVYYNKKK--ALNSVSLDFYPNEITALIGPSGSGKSTLLRSINrmND---------LNPEvtitgSIVYNGHN 73
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VhqnmgYCPQFDAItEL---------------LTGREHVEFFALLRGVPEKEV-GKVGEWAIRKLGL---VK-YGEKYAS 2045
Cdd:PRK14239 74 I-----YSPRTDTV-DLrkeigmvfqqpnpfpMSIYENVVYGLRLKGIKDKQVlDEAVEKSLKGASIwdeVKdRLHDSAL 147
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 2046 NYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIvKEGRSVVLTSHSMEECEALCTRMAIMVNG 2122
Cdd:PRK14239 148 GLSGGQQQRVCIARVLATSPKIILLDEPTSALDPISAGKIEETLLGL-KDDYTMLLVTRSMQQASRISDRTGFFLDG 223
|
|
| PRK15112 |
PRK15112 |
peptide ABC transporter ATP-binding protein SapF; |
901-1115 |
3.55e-08 |
|
peptide ABC transporter ATP-binding protein SapF;
Pssm-ID: 185067 [Multi-domain] Cd Length: 267 Bit Score: 57.11 E-value: 3.55e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 901 IQNLVKV--YRDGM-----KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGK-----DIRSEM 968
Cdd:PRK15112 7 VRNLSKTfrYRTGWfrrqtVEAVKPLSFTLREGQTLAIIGENGSGKSTLAKMLAGMIEPTSGELLIDDHplhfgDYSYRS 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 SSIRQ-----NLGVCPQHNV--LFDM-------LTVEEHiwfyarlkglsEKHVKAEMEQmaldVGLPPSKLKSKTSQLS 1034
Cdd:PRK15112 87 QRIRMifqdpSTSLNPRQRIsqILDFplrlntdLEPEQR-----------EKQIIETLRQ----VGLLPDHASYYPHMLA 151
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1035 GGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKY--RQGRTIILSTHHMDEADILGDRIAIISHGKLCCV 1112
Cdd:PRK15112 152 PGQKQRLGLARALILRPKVIIADEALASLDMSMRSQLINLMLELqeKQGISYIYVTQHLGMMKHISDQVLVMHQGEVVER 231
|
...
gi 568925948 1113 GSS 1115
Cdd:PRK15112 232 GST 234
|
|
| PLN03232 |
PLN03232 |
ABC transporter C family member; Provisional |
870-1109 |
3.67e-08 |
|
ABC transporter C family member; Provisional
Pssm-ID: 215640 [Multi-domain] Cd Length: 1495 Bit Score: 59.22 E-value: 3.67e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 870 IDEKSHPGSSQKGVSEICMEEepTHLRlgvsiqnlvkvYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGL 949
Cdd:PLN03232 1219 IIENNRPVSGWPSRGSIKFED--VHLR-----------YRPGLPPVLHGLSFFVSPSEKVGVVGRTGAGKSSMLNALFRI 1285
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 950 FPPTSGTAYILGKDI-RSEMSSIRQNLGVCPQHNVLF------DMLTVEEH----IWfyarlKGLSEKHVKAEMEQ--MA 1016
Cdd:PLN03232 1286 VELEKGRIMIDDCDVaKFGLTDLRRVLSIIPQSPVLFsgtvrfNIDPFSEHndadLW-----EALERAHIKDVIDRnpFG 1360
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1017 LDvglppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEAdI 1096
Cdd:PLN03232 1361 LD-----AEVSEGGENFSVGQRQLLSLARALLRRSKILVLDEATASVDVRTDSLIQRTIREEFKSCTMLVIAHRLNTI-I 1434
|
250
....*....|...
gi 568925948 1097 LGDRIAIISHGKL 1109
Cdd:PLN03232 1435 DCDKILVLSSGQV 1447
|
|
| MsbA_lipidA |
TIGR02203 |
lipid A export permease/ATP-binding protein MsbA; This family consists of a single polypeptide ... |
1914-2123 |
3.71e-08 |
|
lipid A export permease/ATP-binding protein MsbA; This family consists of a single polypeptide chain transporter in the ATP-binding cassette (ABC) transporter family, MsbA, which exports lipid A. It may also act in multidrug resistance. Lipid A, a part of lipopolysaccharide, is found in the outer leaflet of the outer membrane of most Gram-negative bacteria. Members of this family are restricted to the Proteobacteria (although lipid A is more broadly distributed) and often are clustered with lipid A biosynthesis genes. [Cell envelope, Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides, Transport and binding proteins, Other]
Pssm-ID: 131258 [Multi-domain] Cd Length: 571 Bit Score: 58.57 E-value: 3.71e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNK--------NSILSNIHE 1985
Cdd:TIGR02203 331 VEFRNVTFRYPGRDRPALDSISLVIEPGETVALVGRSGSGKSTLVNLIPRFYEPDSGQILLDGhdladytlASLRRQVAL 410
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYcpqFDAitellTGREHVEFFALlRGVPEKEVGKVGEWAIRK-------LGL-VKYGEKyASNYSGGNKRKLST 2057
Cdd:TIGR02203 411 VSQDVVL---FND-----TIANNIAYGRT-EQADRAEIERALAAAYAQdfvdklpLGLdTPIGEN-GVLLSGGQRQRLAI 480
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRfLWNCALSIVKEGRSVVLTSHSMEECEAlCTRMAIMVNGR 2123
Cdd:TIGR02203 481 ARALLKDAPILILDEATSALDNESER-LVQAALERLMQGRTTLVIAHRLSTIEK-ADRIVVMDDGR 544
|
|
| artP |
PRK11124 |
arginine transporter ATP-binding subunit; Provisional |
1934-2135 |
4.47e-08 |
|
arginine transporter ATP-binding subunit; Provisional
Pssm-ID: 182980 [Multi-domain] Cd Length: 242 Bit Score: 56.18 E-value: 4.47e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1934 ICIGIPPGECFGLLGVNGAGKSTTFKML-------TGDTPVTrGDAF-LNKNSILSNIHEVHQNMGYCPQFDAITELLTG 2005
Cdd:PRK11124 21 ITLDCPQGETLVLLGPSGAGKSSLLRVLnllemprSGTLNIA-GNHFdFSKTPSDKAIRELRRNVGMVFQQYNLWPHLTV 99
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2006 REH-VEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKarrf 2084
Cdd:PRK11124 100 QQNlIEAPCRVLGLSKDQALARAEKLLERLRLKPYADRFPLHLSGGQQQRVAIARALMMEPQVLLFDEPTAALDPE---- 175
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2085 LWNCALSIVKE----GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKN 2135
Cdd:PRK11124 176 ITAQIVSIIRElaetGITQVIVTHEVEVARKTASRVVYMENGHIVEQGDASCFTQ 230
|
|
| PRK13541 |
PRK13541 |
cytochrome c biogenesis protein CcmA; Provisional |
919-1089 |
5.24e-08 |
|
cytochrome c biogenesis protein CcmA; Provisional
Pssm-ID: 184128 [Multi-domain] Cd Length: 195 Bit Score: 55.26 E-value: 5.24e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 919 LALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI----RSEMSSIRQNLGVCPQhnvlfdmLTVEEHI 994
Cdd:PRK13541 19 LSITFLPSAITYIKGANGCGKSSLLRMIAGIMQPSSGNIYYKNCNInniaKPYCTYIGHNLGLKLE-------MTVFENL 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 995 WFYARLKGLSEKhVKAEMEQMALDvglppSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWEL 1074
Cdd:PRK13541 92 KFWSEIYNSAET-LYAAIHYFKLH-----DLLDEKCYSLSSGMQKIVAIARLIACQSDLWLLDEVETNLSKENRDLLNNL 165
|
170
....*....|....*.
gi 568925948 1075 L-LKYRQGRTIILSTH 1089
Cdd:PRK13541 166 IvMKANSGGIVLLSSH 181
|
|
| MdlB |
COG1132 |
ABC-type multidrug transport system, ATPase and permease component [Defense mechanisms]; |
1928-2104 |
6.71e-08 |
|
ABC-type multidrug transport system, ATPase and permease component [Defense mechanisms];
Pssm-ID: 440747 [Multi-domain] Cd Length: 579 Bit Score: 57.87 E-value: 6.71e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQfDAIteLLTG- 2005
Cdd:COG1132 353 RPVLKDISLTIPPGETVALVGPSGSGKSTLVNLLLRFYDPTSGRILIDGVDIRDlTLESLRRQIGVVPQ-DTF--LFSGt 429
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2006 -REHVEFFALlrGVPEKEVgkvgEWAIRKLGLVKY------------GEKyASNYSGGNKRKLSTAMALIGGPPVVFLDE 2072
Cdd:COG1132 430 iRENIRYGRP--DATDEEV----EEAAKAAQAHEFiealpdgydtvvGER-GVNLSGGQRQRIAIARALLKDPPILILDE 502
|
170 180 190
....*....|....*....|....*....|..
gi 568925948 2073 PTTGMDPKARRFLWNcALSIVKEGRSVVLTSH 2104
Cdd:COG1132 503 ATSALDTETEALIQE-ALERLMKGRTTIVIAH 533
|
|
| modC_ABC |
TIGR02142 |
molybdenum ABC transporter, ATP-binding protein; This model represents the ATP-binding ... |
1938-2136 |
7.38e-08 |
|
molybdenum ABC transporter, ATP-binding protein; This model represents the ATP-binding cassette (ABC) protein of the three subunit molybdate ABC transporter. The three proteins of this complex are homologous to proteins of the sulfate ABC transporter. Molybdenum may be used in nitrogenases of nitrogen-fixing bacteria and in molybdopterin cofactors. In some cases, molybdate may be transported by a sulfate transporter rather than by a specific molybdate transporter. [Transport and binding proteins, Anions]
Pssm-ID: 131197 [Multi-domain] Cd Length: 354 Bit Score: 57.04 E-value: 7.38e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLN--------KNSILSnIHE-----VHQNMGYCPQFDAITELLT 2004
Cdd:TIGR02142 20 LPGQGVTAIFGRSGSGKTTLIRLIAGLTRPDEGEIVLNgrtlfdsrKGIFLP-PEKrrigyVFQEARLFPHLSVRGNLRY 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2005 GREHVEffALLRGVPEKEVgkvgewaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARR- 2083
Cdd:TIGR02142 99 GMKRAR--PSERRISFERV-------IELLGIGHLLGRLPGRLSGGEKQRVAIGRALLSSPRLLLMDEPLAALDDPRKYe 169
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 2084 ---FLWNCALSIvkeGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNR 2136
Cdd:TIGR02142 170 ilpYLERLHAEF---GIPILYVSHSLQEVLRLADRVVVLEDGRVAAAGPIAEVWAS 222
|
|
| ABCC_ATM1_transporter |
cd03253 |
ATP-binding cassette domain of iron-sulfur clusters transporter, subfamily C; ATM1 is an ABC ... |
1928-2104 |
7.41e-08 |
|
ATP-binding cassette domain of iron-sulfur clusters transporter, subfamily C; ATM1 is an ABC transporter that is expressed in the mitochondria. Although the specific function of ATM1 is unknown, its disruption results in the accumulation of excess mitochondrial iron, loss of mitochondrial cytochromes, oxidative damage to mitochondrial DNA, and decreased levels of cytosolic heme proteins. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins.
Pssm-ID: 213220 [Multi-domain] Cd Length: 236 Bit Score: 55.70 E-value: 7.41e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknSIL---SNIHEVHQN-----MGYCPQ---- 1995
Cdd:cd03253 14 RPVLKDVSFTIPAGKKVAIVGPSGSGKSTILRLLFRFYDVSSG-------SILidgQDIREVTLDslrraIGVVPQdtvl 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1996 FDAITE--LLTGR-----EHVEFFA-------LLRGVPEKEVGKVGEwaiRKLGLvkygekyasnySGGNKRKLSTAMAL 2061
Cdd:cd03253 87 FNDTIGynIRYGRpdatdEEVIEAAkaaqihdKIMRFPDGYDTIVGE---RGLKL-----------SGGEKQRVAIARAI 152
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 2062 IGGPPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSH 2104
Cdd:cd03253 153 LKNPPILLLDEATSALDTHTEREIQA-ALRDVSKGRTTIVIAH 194
|
|
| PRK13543 |
PRK13543 |
heme ABC exporter ATP-binding protein CcmA; |
1923-2116 |
8.25e-08 |
|
heme ABC exporter ATP-binding protein CcmA;
Pssm-ID: 184129 [Multi-domain] Cd Length: 214 Bit Score: 55.24 E-value: 8.25e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1923 YRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNihEVHQNMGYCPQFDAITEL 2002
Cdd:PRK13543 19 FSRNEEPVFGPLDFHVDAGEALLVQGDNGAGKTTLLRVLAGLLHVESGQIQIDGKTATRG--DRSRFMAYLGHLPGLKAD 96
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2003 LTGREHVEFFALLRGV-PEKEVGKvgewAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKA 2081
Cdd:PRK13543 97 LSTLENLHFLCGLHGRrAKQMPGS----ALAIVGLAGYEDTLVRQLSAGQKKRLALARLWLSPAPLWLLDEPYANLDLEG 172
|
170 180 190
....*....|....*....|....*....|....*
gi 568925948 2082 RRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRM 2116
Cdd:PRK13543 173 ITLVNRMISAHLRGGGAALVTTHGAYAAPPVRTRM 207
|
|
| PRK13549 |
PRK13549 |
xylose transporter ATP-binding subunit; Provisional |
1913-2123 |
9.67e-08 |
|
xylose transporter ATP-binding subunit; Provisional
Pssm-ID: 184134 [Multi-domain] Cd Length: 506 Bit Score: 57.25 E-value: 9.67e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVT--RGDAFLNKNSI-LSNIHE---- 1985
Cdd:PRK13549 5 LLEMKNITKTFGGVK--ALDNVSLKVRAGEIVSLCGENGAGKSTLMKVLSGVYPHGtyEGEIIFEGEELqASNIRDtera 82
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 ----VHQNMGYCPQFDAITELLTGREHVEFfallrGVP--EKEVGKVGEWaIRKLGLVKYGEKYASNYSGGNKRKLSTAM 2059
Cdd:PRK13549 83 giaiIHQELALVKELSVLENIFLGNEITPG-----GIMdyDAMYLRAQKL-LAQLKLDINPATPVGNLGLGQQQLVEIAK 156
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*...
gi 568925948 2060 ALIGGPPVVFLDEPTTGMDPKARRFLwncaLSIVKE----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK13549 157 ALNKQARLLILDEPTASLTESETAVL----LDIIRDlkahGIACIYISHKLNEVKAISDTICVIRDGR 220
|
|
| ssuB |
PRK11247 |
aliphatic sulfonates transport ATP-binding subunit; Provisional |
1914-2123 |
9.70e-08 |
|
aliphatic sulfonates transport ATP-binding subunit; Provisional
Pssm-ID: 183055 [Multi-domain] Cd Length: 257 Bit Score: 55.45 E-value: 9.70e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRkpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPvTRGDaFLNKNSILSNIHEVHQNMgy 1992
Cdd:PRK11247 13 LLLNAVSKRYGERT--VLNQLDLHIPAGQFVAVVGRSGCGKSTLLRLLAGlETP-SAGE-LLAGTAPLAEAREDTRLM-- 86
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 cpqfdaitelltgrehvefFALLRGVPEKEV------GKVGEW------AIRKLGLVKYGEKYASNYSGGNKRKLSTAMA 2060
Cdd:PRK11247 87 -------------------FQDARLLPWKKVidnvglGLKGQWrdaalqALAAVGLADRANEWPAALSGGQKQRVALARA 147
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2061 LIGGPPVVFLDEPTTGMDPKAR-------RFLWNcalsivKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK11247 148 LIHRPGLLLLDEPLGALDALTRiemqdliESLWQ------QHGFTVLLVTHDVSEAVAMADRVLLIEEGK 211
|
|
| PRK10261 |
PRK10261 |
glutathione transporter ATP-binding protein; Provisional |
915-1113 |
9.86e-08 |
|
glutathione transporter ATP-binding protein; Provisional
Pssm-ID: 182342 [Multi-domain] Cd Length: 623 Bit Score: 57.17 E-value: 9.86e-08
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIR----SEMSSIRQNLGVCPQ--HNVLFDML 988
Cdd:PRK10261 339 AVEKVSFDLWPGETLSLVGESGSGKSTTGRALLRLVESQGGEIIFNGQRIDtlspGKLQALRRDIQFIFQdpYASLDPRQ 418
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWFYARLKGLSEKHVKAEMEQMALD-VGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYS 1067
Cdd:PRK10261 419 TVGDSIMEPLRVHGLLPGKAAAARVAWLLErVGLLPEHAWRYPHEFSGGQRQRICIARALALNPKVIIADEAVSALDVSI 498
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1068 RRGIWELLLKYRQ--GRTIILSTHHMDEADILGDRIAIISHGKLCCVG 1113
Cdd:PRK10261 499 RGQIINLLLDLQRdfGIAYLFISHDMAVVERISHRVAVMYLGQIVEIG 546
|
|
| ABC_Class2 |
cd03227 |
ATP-binding cassette domain of non-transporter proteins; ABC-type Class 2 contains systems ... |
1032-1089 |
1.10e-07 |
|
ATP-binding cassette domain of non-transporter proteins; ABC-type Class 2 contains systems involved in cellular processes other than transport. These families are characterized by the fact that the ABC subunit is made up of duplicated, fused ABC modules (ABC2). No known transmembrane proteins or domains are associated with these proteins.
Pssm-ID: 213194 [Multi-domain] Cd Length: 162 Bit Score: 53.52 E-value: 1.10e-07
10 20 30 40 50 60
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1032 QLSGGMQRKLSVALAF----VGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ-GRTIILSTH 1089
Cdd:cd03227 77 QLSGGEKELSALALILalasLKPRPLYILDEIDRGLDPRDGQALAEAILEHLVkGAQVIVITH 139
|
|
| PRK11176 |
PRK11176 |
lipid A ABC transporter ATP-binding protein/permease MsbA; |
1914-2104 |
1.24e-07 |
|
lipid A ABC transporter ATP-binding protein/permease MsbA;
Pssm-ID: 183016 [Multi-domain] Cd Length: 582 Bit Score: 56.95 E-value: 1.24e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI----LSNIHE---- 1985
Cdd:PRK11176 342 IEFRNVTFTYPGKEVPALRNINFKIPAGKTVALVGRSGSGKSTIANLLTRFYDIDEGEILLDGHDLrdytLASLRNqval 421
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 ----VH-------QNMGYcpqfdAITELLTgREHVEFFALLRGVPEkevgkvgewAIRKL--GL-VKYGEKYASnYSGGN 2051
Cdd:PRK11176 422 vsqnVHlfndtiaNNIAY-----ARTEQYS-REQIEEAARMAYAMD---------FINKMdnGLdTVIGENGVL-LSGGQ 485
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2052 KRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLwNCALSIVKEGRSVVLTSH 2104
Cdd:PRK11176 486 RQRIAIARALLRDSPILILDEATSALDTESERAI-QAALDELQKNRTSLVIAH 537
|
|
| CFTR_protein |
TIGR01271 |
cystic fibrosis transmembrane conductor regulator (CFTR); The model describes the cystis ... |
899-1092 |
1.39e-07 |
|
cystic fibrosis transmembrane conductor regulator (CFTR); The model describes the cystis fibrosis transmembrane conductor regulator (CFTR) in eukaryotes. The principal role of this protein is chloride ion conductance. The protein is predicted to consist of 12 transmembrane domains. Mutations or lesions in the genetic loci have been linked to the aetiology of asthma, bronchiectasis, chronic obstructive pulmonary disease etc. Disease-causing mutations have been studied by 36Cl efflux assays in vitro cell cultures and electrophysiology, all of which point to the impairment of chloride channel stability and not the biosynthetic processing per se. [Transport and binding proteins, Anions]
Pssm-ID: 273530 [Multi-domain] Cd Length: 1490 Bit Score: 57.23 E-value: 1.39e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFpPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:TIGR01271 1218 MDVQGLTAKYTEAGRAVLQDLSFSVEGGQRVGLLGRTGSGKSTLLSALLRLL-STEGEIQIDGVSWNSvTLQTWRKAFGV 1296
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFD----------MLTVEEHIWFYARLKGLsekhvKAEMEQMaldvglpPSKLKSKTSQ----LSGGMQRKLSV 1043
Cdd:TIGR01271 1297 IPQKVFIFSgtfrknldpyEQWSDEEIWKVAEEVGL-----KSVIEQF-------PDKLDFVLVDggyvLSNGHKQLMCL 1364
|
170 180 190 200
....*....|....*....|....*....|....*....|....*....
gi 568925948 1044 ALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMD 1092
Cdd:TIGR01271 1365 ARSILSKAKILLLDEPSAHLDPVTLQIIRKTLKQSFSNCTVILSEHRVE 1413
|
|
| ABCC_SUR1_N |
cd03290 |
ATP-binding cassette domain of the sulfonylurea receptor, subfamily C; The SUR domain 1. The ... |
926-1091 |
1.42e-07 |
|
ATP-binding cassette domain of the sulfonylurea receptor, subfamily C; The SUR domain 1. The sulfonylurea receptor SUR is an ATP transporter of the ABCC/MRP family with tandem ATPase binding domains. Unlike other ABC proteins, it has no intrinsic transport function, neither active nor passive, but associates with the potassium channel proteins Kir6.1 or Kir6.2 to form the ATP-sensitive potassium (K(ATP)) channel. Within the channel complex, SUR serves as a regulatory subunit that fine-tunes the gating of Kir6.x in response to alterations in cellular metabolism. It constitutes a major pharmaceutical target as it binds numerous drugs, K(ATP) channel openers and blockers, capable of up- or down-regulating channel activity.
Pssm-ID: 213257 [Multi-domain] Cd Length: 218 Bit Score: 54.64 E-value: 1.42e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSI-----RQNLGVCPQHNVLFDMlTVEEHIWFYARL 1000
Cdd:cd03290 27 GQLTMIVGQVGCGKSSLLLAILGEMQTLEGKVHWSNKNESEPSFEAtrsrnRYSVAYAAQKPWLLNA-TVEENITFGSPF 105
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1001 kglSEKHVKAEMEQMAL--DVGLPP----SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPY-SRRGIWE 1073
Cdd:cd03290 106 ---NKQRYKAVTDACSLqpDIDLLPfgdqTEIGERGINLSGGQRQRICVARALYQNTNIVFLDDPFSALDIHlSDHLMQE 182
|
170 180
....*....|....*....|
gi 568925948 1074 LLLKYRQG--RTIILSTHHM 1091
Cdd:cd03290 183 GILKFLQDdkRTLVLVTHKL 202
|
|
| PRK10636 |
PRK10636 |
putative ABC transporter ATP-binding protein; Provisional |
911-1087 |
1.68e-07 |
|
putative ABC transporter ATP-binding protein; Provisional
Pssm-ID: 236729 [Multi-domain] Cd Length: 638 Bit Score: 56.72 E-value: 1.68e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYiLGKDIRsemssirqnLGVCPQHNVLFdMLTV 990
Cdd:PRK10636 323 GDRIILDSIKLNLVPGSRIGLLGRNGAGKSTLIKLLAGELAPVSGEIG-LAKGIK---------LGYFAQHQLEF-LRAD 391
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 991 EEHIWFYARLkglsekhVKAEMEQMALD----VGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPY 1066
Cdd:PRK10636 392 ESPLQHLARL-------APQELEQKLRDylggFGFQGDKVTEETRRFSGGEKARLVLALIVWQRPNLLLLDEPTNHLDLD 464
|
170 180
....*....|....*....|.
gi 568925948 1067 SRRGIWELLLKYrQGRTIILS 1087
Cdd:PRK10636 465 MRQALTEALIDF-EGALVVVS 484
|
|
| ABCC_CFTR2 |
cd03289 |
ATP-binding cassette domain 2 of CFTR,subfamily C; The cystic fibrosis transmembrane regulator ... |
899-1109 |
2.59e-07 |
|
ATP-binding cassette domain 2 of CFTR,subfamily C; The cystic fibrosis transmembrane regulator (CFTR), the product of the gene mutated in patients with cystic fibrosis, has adapted the ABC transporter structural motif to form a tightly regulated anion channel at the apical surface of many epithelia. Use of the term assembly of a functional ion channel implies the coming together of subunits or at least smaller not-yet functional components of the active whole. In fact, on the basis of current knowledge only the CFTR polypeptide itself is required to form an ATP- and protein kinase A-dependent low-conductance chloride channel of the type present in the apical membrane of many epithelial cells. CFTR displays the typical organization (IM-ABC)2 and carries a characteristic hydrophilic R-domain that separates IM1-ABC1 from IM2-ABC2.
Pssm-ID: 213256 [Multi-domain] Cd Length: 275 Bit Score: 54.48 E-value: 2.59e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPpTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:cd03289 3 MTVKDLTAKYTEGGNAVLENISFSISPGQRVGLLGRTGSGKSTLLSAFLRLLN-TEGDIQIDGVSWNSvPLQKWRKAFGV 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMlTVEEHIWFYarlkglsEKHVKAEMEQMALDVGLP------PSKLKSKTSQ----LSGGMQRKLSVALAF 1047
Cdd:cd03289 82 IPQKVFIFSG-TFRKNLDPY-------GKWSDEEIWKVAEEVGLKsvieqfPGQLDFVLVDggcvLSHGHKQLMCLARSV 153
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMdEADILGDRIAIISHGKL 1109
Cdd:cd03289 154 LSKAKILLLDEPSAHLDPITYQVIRKTLKQAFADCTVILSEHRI-EAMLECQRFLVIEENKV 214
|
|
| ABC_ABC_ChvD |
TIGR03719 |
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of ... |
896-1087 |
3.66e-07 |
|
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of the ABC transporter ATP-binding cassette, but are found outside the common ABC transporter operon structure that features integral membrane permease proteins and substrate-binding proteins encoded next to the ATP-binding cassette (ABC domain) protein. The member protein ChvD from Agrobacterium tumefaciens was identified as both a candidate to interact with VirB8, based on yeast two-hybrid analysis, and as an apparent regulator of VirG. The general function of this protein family is unknown.
Pssm-ID: 274744 [Multi-domain] Cd Length: 552 Bit Score: 55.33 E-value: 3.66e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 896 RLG---VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTaYILGKDIRseMSSIR 972
Cdd:TIGR03719 317 RLGdkvIEAENLTKAFGD--KLLIDDLSFKLPPGGIVGVIGPNGAGKSTLFRMITGQEQPDSGT-IEIGETVK--LAYVD 391
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNlgvcpqHNVLFDMLTVEEHIwfyarLKGLSEKHV-KAEMEQMALdVG---LPPSKLKSKTSQLSGGMQRKLSVALAFV 1048
Cdd:TIGR03719 392 QS------RDALDPNKTVWEEI-----SGGLDIIKLgKREIPSRAY-VGrfnFKGSDQQKKVGQLSGGERNRVHLAKTLK 459
|
170 180 190
....*....|....*....|....*....|....*....
gi 568925948 1049 GGSKVVILDEPTAGVDPYSRRGIWELLLKYrQGRTIILS 1087
Cdd:TIGR03719 460 SGGNVLLLDEPTNDLDVETLRALEEALLNF-AGCAVVIS 497
|
|
| PRK13549 |
PRK13549 |
xylose transporter ATP-binding subunit; Provisional |
1913-2125 |
4.02e-07 |
|
xylose transporter ATP-binding subunit; Provisional
Pssm-ID: 184134 [Multi-domain] Cd Length: 506 Bit Score: 55.32 E-value: 4.02e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIY----RRKRkpaVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP-VTRGDAFLN--KNSILSNIHE 1985
Cdd:PRK13549 259 ILEVRNLTAWDpvnpHIKR---VDDVSFSLRRGEILGIAGLVGAGRTELVQCLFGAYPgRWEGEIFIDgkPVKIRNPQQA 335
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYCPQ---FDAITELLTGREHVEFFAL----LRGV--PEKEVGKVGEwAIRKLGLvkygeKYAS------NYSGG 2050
Cdd:PRK13549 336 IAQGIAMVPEdrkRDGIVPVMGVGKNITLAALdrftGGSRidDAAELKTILE-SIQRLKV-----KTASpelaiaRLSGG 409
|
170 180 190 200 210 220 230
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2051 NKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGRFR 2125
Cdd:PRK13549 410 NQQKAVLAKCLLLNPKILILDEPTRGIDVGAKYEIYKLINQLVQQGVAIIVISSELPEVLGLSDRVLVMHEGKLK 484
|
|
| PRK10790 |
PRK10790 |
SmdB family multidrug efflux ABC transporter permease/ATP-binding protein; |
1914-2104 |
4.52e-07 |
|
SmdB family multidrug efflux ABC transporter permease/ATP-binding protein;
Pssm-ID: 182733 [Multi-domain] Cd Length: 592 Bit Score: 55.11 E-value: 4.52e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKrKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEV-HQNMGY 1992
Cdd:PRK10790 341 IDIDNVSFAYRDD-NLVLQNINLSVPSRGFVALVGHTGSGKSTLASLLMGYYPLTEGEIRLDGRPLSSLSHSVlRQGVAM 419
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1993 CPQ---------FDAITellTGR-----------EHVEFFALLRGVPEKEVGKVGEwairklglvkygekYASNYSGGNK 2052
Cdd:PRK10790 420 VQQdpvvladtfLANVT---LGRdiseeqvwqalETVQLAELARSLPDGLYTPLGE--------------QGNNLSVGQK 482
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2053 RKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNcALSIVKEGRSVVLTSH 2104
Cdd:PRK10790 483 QLLALARVLVQTPQILILDEATANIDSGTEQAIQQ-ALAAVREHTTLVVIAH 533
|
|
| znuC |
PRK09544 |
high-affinity zinc transporter ATPase; Reviewed |
911-1114 |
5.05e-07 |
|
high-affinity zinc transporter ATPase; Reviewed
Pssm-ID: 181939 [Multi-domain] Cd Length: 251 Bit Score: 53.19 E-value: 5.05e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayilgkdIRSEmSSIRqnLGVCPQhNVLFDM--- 987
Cdd:PRK09544 15 GQRRVLSDVSLELKPGKILTLLGPNGAGKSTLVRVVLGLVAPDEGV-------IKRN-GKLR--IGYVPQ-KLYLDTtlp 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 988 LTVEEhiwfYARLKGlsekHVKAEMEQMALDVGLPPSKLKSKTSQLSGG-MQRKLsVALAFVGGSKVVILDEPTAGVDPY 1066
Cdd:PRK09544 84 LTVNR----FLRLRP----GTKKEDILPALKRVQAGHLIDAPMQKLSGGeTQRVL-LARALLNRPQLLVLDEPTQGVDVN 154
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1067 SRRGIWELLLKYRQ--GRTIILSTH--HMDEADIlgDRIAIISHgKLCCVGS 1114
Cdd:PRK09544 155 GQVALYDLIDQLRRelDCAVLMVSHdlHLVMAKT--DEVLCLNH-HICCSGT 203
|
|
| PRK10908 |
PRK10908 |
cell division ATP-binding protein FtsE; |
1913-2123 |
5.45e-07 |
|
cell division ATP-binding protein FtsE;
Pssm-ID: 182829 [Multi-domain] Cd Length: 222 Bit Score: 52.95 E-value: 5.45e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIL----SNIHEVHQ 1988
Cdd:PRK10908 1 MIRFEHVSKAYLGGRQ-ALQGVTFHMRPGEMAFLTGHSGAGKSTLLKLICGIERPSAGKIWFSGHDITrlknREVPFLRR 79
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVV 2068
Cdd:PRK10908 80 QIGMIFQDHHLLMDRTVYDNVAIPLIIAGASGDDIRRRVSAALDKVGLLDKAKNFPIQLSGGEQQRVGIARAVVNKPAVL 159
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2069 FLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:PRK10908 160 LADEPTGNLDDALSEGILRLFEEFNRVGVTVLMATHDIGLISRRSYRMLTLSDGH 214
|
|
| 3a01205 |
TIGR00956 |
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other] |
1938-2078 |
5.68e-07 |
|
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273362 [Multi-domain] Cd Length: 1394 Bit Score: 55.11 E-value: 5.68e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTpvtrgDAFL-NKNSILS-NIHEVHQNMG-------YCPQFDAITELLTGREH 2008
Cdd:TIGR00956 84 IKPGELTVVLGRPGSGCSTLLKTIASNT-----DGFHiGVEGVITyDGITPEEIKKhyrgdvvYNAETDVHFPHLTVGET 158
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2009 VEFFALLR-------GVPEKE-VGKVGEWAIRKLGL-----VKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTT 2075
Cdd:TIGR00956 159 LDFAARCKtpqnrpdGVSREEyAKHIADVYMATYGLshtrnTKVGNDFVRGVSGGERKRVSIAEASLGGAKIQCWDNATR 238
|
...
gi 568925948 2076 GMD 2078
Cdd:TIGR00956 239 GLD 241
|
|
| PRK11831 |
PRK11831 |
phospholipid ABC transporter ATP-binding protein MlaF; |
1910-2135 |
7.83e-07 |
|
phospholipid ABC transporter ATP-binding protein MlaF;
Pssm-ID: 236997 [Multi-domain] Cd Length: 269 Bit Score: 52.85 E-value: 7.83e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTkiYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSI----LSNIHE 1985
Cdd:PRK11831 4 VANLVDMRGVS--FTRGNRCIFDNISLTVPRGKITAIMGPSGIGKTTLLRLIGGQIAPDHGEILFDGENIpamsRSRLYT 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQNMGYCPQFDAITELLTGREHVEFfallrgvPEKEVGKVGEWAIR-----KL---GLVKYGEKYASNYSGGNKRKLST 2057
Cdd:PRK11831 82 VRKRMSMLFQSGALFTDMNVFDNVAY-------PLREHTQLPAPLLHstvmmKLeavGLRGAAKLMPSELSGGMARRAAL 154
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2058 AMALIGGPPVVFLDEPTTGMDPKARRFL------WNCALsivkeGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQ 2131
Cdd:PRK11831 155 ARAIALEPDLIMFDEPFVGQDPITMGVLvkliseLNSAL-----GVTCVVVSHDVPEVLSIADHAYIVADKKIVAHGSAQ 229
|
....
gi 568925948 2132 HLKN 2135
Cdd:PRK11831 230 ALQA 233
|
|
| ABCC_MRP_domain2 |
cd03244 |
ATP-binding cassette domain 2 of multidrug resistance-associated protein; The ABC subfamily C ... |
1914-2079 |
9.50e-07 |
|
ATP-binding cassette domain 2 of multidrug resistance-associated protein; The ABC subfamily C is also known as MRP (multidrug resistance-associated protein). Some of the MRP members have five additional transmembrane segments in their N-terminus, but the function of these additional membrane-spanning domains is not clear. The MRP was found in the multidrug-resistance lung cancer cell in which p-glycoprotein was not overexpressed. MRP exports glutathione by drug stimulation, as well as, certain substrates in conjugated forms with anions, such as glutathione, glucuronate, and sulfate.
Pssm-ID: 213211 [Multi-domain] Cd Length: 221 Bit Score: 52.11 E-value: 9.50e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTT----FKMLtgdtPVTRGDAFLNKNSILS-NIHEVHQ 1988
Cdd:cd03244 3 IEFKNVSLRYRPNLPPVLKNISFSIKPGEKVGIVGRTGSGKSSLllalFRLV----ELSSGSILIDGVDISKiGLHDLRS 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1989 NMGYCPQfDAIteLLTG--REHVEFFALlrgVPEKEVgkvgeW-AIRKLGLVKYGEKYA-----------SNYSGGNKRK 2054
Cdd:cd03244 79 RISIIPQ-DPV--LFSGtiRSNLDPFGE---YSDEEL-----WqALERVGLKEFVESLPggldtvveeggENLSVGQRQL 147
|
170 180
....*....|....*....|....*
gi 568925948 2055 LSTAMALIGGPPVVFLDEPTTGMDP 2079
Cdd:cd03244 148 LCLARALLRKSKILVLDEATASVDP 172
|
|
| PRK13540 |
PRK13540 |
cytochrome c biogenesis protein CcmA; Provisional |
1928-2104 |
9.60e-07 |
|
cytochrome c biogenesis protein CcmA; Provisional
Pssm-ID: 184127 [Multi-domain] Cd Length: 200 Bit Score: 51.49 E-value: 9.60e-07
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1928 KPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVHQNMGYCPQFDAITELLTGRE 2007
Cdd:PRK13540 14 QPLLQQISFHLPAGGLLHLKGSNGAGKTTLLKLIAGLLNPEKGEILFERQSIKKDLCTYQKQLCFVGHRSGINPYLTLRE 93
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 HVeFFALLRGVPEKEVGKVgewaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWN 2087
Cdd:PRK13540 94 NC-LYDIHFSPGAVGITEL----CRLFSLEHLIDYPCGLLSSGQKRQVALLRLWMSKAKLWLLDEPLVALDELSLLTIIT 168
|
170
....*....|....*..
gi 568925948 2088 CALSIVKEGRSVVLTSH 2104
Cdd:PRK13540 169 KIQEHRAKGGAVLLTSH 185
|
|
| PRK13543 |
PRK13543 |
heme ABC exporter ATP-binding protein CcmA; |
933-1065 |
1.04e-06 |
|
heme ABC exporter ATP-binding protein CcmA;
Pssm-ID: 184129 [Multi-domain] Cd Length: 214 Bit Score: 51.77 E-value: 1.04e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 933 GHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-RSEMSSIRQNLGVCPQhnvLFDMLTVEEHIWFyarLKGLSEKHVKaE 1011
Cdd:PRK13543 44 GDNGAGKTTLLRVLAGLLHVESGQIQIDGKTAtRGDRSRFMAYLGHLPG---LKADLSTLENLHF---LCGLHGRRAK-Q 116
|
90 100 110 120 130
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1012 MEQMALD-VGLPPSKlKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDP 1065
Cdd:PRK13543 117 MPGSALAiVGLAGYE-DTLVRQLSAGQKKRLALARLWLSPAPLWLLDEPYANLDL 170
|
|
| oppD |
PRK09473 |
oligopeptide transporter ATP-binding component; Provisional |
1930-2133 |
1.09e-06 |
|
oligopeptide transporter ATP-binding component; Provisional
Pssm-ID: 181888 [Multi-domain] Cd Length: 330 Bit Score: 53.19 E-value: 1.09e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG---DTPVTRGDAFLNKNSILsNIHEVHQNMGYCPQF-----DAITE 2001
Cdd:PRK09473 31 AVNDLNFSLRAGETLGIVGESGSGKSQTAFALMGllaANGRIGGSATFNGREIL-NLPEKELNKLRAEQIsmifqDPMTS 109
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2002 L----LTGREHVEFFALLRGVPEKEVGkvgEWAIRKLGLVKYGEK------YASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:PRK09473 110 LnpymRVGEQLMEVLMLHKGMSKAEAF---EESVRMLDAVKMPEArkrmkmYPHEFSGGMRQRVMIAMALLCRPKLLIAD 186
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 2072 EPTTGMDPKARRFLWNCALSIVKE-GRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK09473 187 EPTTALDVTVQAQIMTLLNELKREfNTAIIMITHDLGVVAGICDKVLVMYAGRTMEYGNARDV 249
|
|
| dppD |
PRK11022 |
dipeptide transporter ATP-binding subunit; Provisional |
915-1089 |
1.16e-06 |
|
dipeptide transporter ATP-binding subunit; Provisional
Pssm-ID: 182906 [Multi-domain] Cd Length: 326 Bit Score: 52.82 E-value: 1.16e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 915 AVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP-PTSGTAYIL---GKDIR------------SEMSSIRQN--LG 976
Cdd:PRK11022 22 AVDRISYSVKQGEVVGIVGESGSGKSVSSLAIMGLIDyPGRVMAEKLefnGQDLQrisekerrnlvgAEVAMIFQDpmTS 101
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 977 VCPQHNVLFD-MLTVEEHiwfyarlKGLSekhvKAEMEQMALD----VGLP--PSKLKSKTSQLSGGMQRKLSVALAFVG 1049
Cdd:PRK11022 102 LNPCYTVGFQiMEAIKVH-------QGGN----KKTRRQRAIDllnqVGIPdpASRLDVYPHQLSGGMSQRVMIAMAIAC 170
|
170 180 190 200
....*....|....*....|....*....|....*....|..
gi 568925948 1050 GSKVVILDEPTAGVDPYSRRGIWELLLKYRQGR--TIILSTH 1089
Cdd:PRK11022 171 RPKLLIADEPTTALDVTIQAQIIELLLELQQKEnmALVLITH 212
|
|
| AAA_21 |
pfam13304 |
AAA domain, putative AbiEii toxin, Type IV TA system; Several members are annotated as being ... |
1775-2107 |
1.26e-06 |
|
AAA domain, putative AbiEii toxin, Type IV TA system; Several members are annotated as being of the abortive phage resistance system, in which case the family would be acting as the toxin for a type IV toxin-antitoxin resistance system.
Pssm-ID: 433102 [Multi-domain] Cd Length: 303 Bit Score: 52.78 E-value: 1.26e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1775 VNLFIGINGSVATFVLELFtnnklndindilksvflifphfclgRGLIDMVKNQAMADALERFGENRFVSPLSWDLvGRN 1854
Cdd:pfam13304 1 INVLIGPNGSGKSNLLEAL-------------------------RFLADFDALVIGLTDERSRNGGIGGIPSLLNG-IDP 54
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1855 LFAMAVEGVVFFLITVLIQYRFFIRPRPVKAKLPPLNDEDEDvRRERQRILDGGGQNDILEIKELTKIYRRKRKPAVDRI 1934
Cdd:pfam13304 55 KEPIEFEISEFLEDGVRYRYGLDLEREDVEEKLSSKPTLLEK-RLLLREDSEEREPKFPPEAEELRLGLDVEERIELSLS 133
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1935 CIGIPpgECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNihevhqnmgycPQFDAITELLTGREHVEFFAL 2014
Cdd:pfam13304 134 ELSDL--ISGLLLLSIISPLSFLLLLDEGLLLEDWAVLDLAADLALFP-----------DLKELLQRLVRGLKLADLNLS 200
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2015 LRGVPEKEVGKVGE-WAIRKLGLVKYGEKY---ASNYSGGNKRKLSTAMALI---GGPPVVFLDEPTTGMDPKARRFLWN 2087
Cdd:pfam13304 201 DLGEGIEKSLLVDDrLRERGLILLENGGGGelpAFELSDGTKRLLALLAALLsalPKGGLLLIDEPESGLHPKLLRRLLE 280
|
330 340
....*....|....*....|
gi 568925948 2088 CALSIVKEGRSVVLTSHSME 2107
Cdd:pfam13304 281 LLKELSRNGAQLILTTHSPL 300
|
|
| ABC_Rad50 |
cd03240 |
ATP-binding cassette domain of Rad50; The catalytic domains of Rad50 are similar to the ... |
923-1090 |
1.57e-06 |
|
ATP-binding cassette domain of Rad50; The catalytic domains of Rad50 are similar to the ATP-binding cassette of ABC transporters, but are not associated with membrane-spanning domains. The conserved ATP-binding motifs common to Rad50 and the ABC transporter family include the Walker A and Walker B motifs, the Q loop, a histidine residue in the switch region, a D-loop, and a conserved LSGG sequence. This conserved sequence, LSGG, is the most specific and characteristic motif of this family and is thus known as the ABC signature sequence.
Pssm-ID: 213207 [Multi-domain] Cd Length: 204 Bit Score: 51.07 E-value: 1.57e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 923 FYEGqITSFLGHNGAGKTTTMS----ILTGLFPPTSGTAYILGKDIRSEMSS----------------IRQNLGV----- 977
Cdd:cd03240 20 FFSP-LTLIVGQNGAGKTTIIEalkyALTGELPPNSKGGAHDPKLIREGEVRaqvklafenangkkytITRSLAIlenvi 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 -CPQhnvlfdmltvEEHIWFYARLKGlsekhvkaemeqmaldvglppsklksktsQLSGGMQRKLSV----ALAFVGGSK 1052
Cdd:cd03240 99 fCHQ----------GESNWPLLDMRG-----------------------------RCSGGEKVLASLiirlALAETFGSN 139
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 1053 --VVILDEPTAGVDPYSRRGIWELLLKYRQG---RTIILSTHH 1090
Cdd:cd03240 140 cgILALDEPTTNLDEENIEESLAEIIEERKSqknFQLIVITHD 182
|
|
| ugpC |
PRK11650 |
sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC; |
898-1107 |
1.74e-06 |
|
sn-glycerol-3-phosphate ABC transporter ATP-binding protein UgpC;
Pssm-ID: 236947 [Multi-domain] Cd Length: 356 Bit Score: 52.54 E-value: 1.74e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 898 GVSIQNLVKVYrDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSEMSSIR----- 972
Cdd:PRK11650 3 GLKLQAVRKSY-DGKTQVIKGIDLDVADGEFIVLVGPSGCGKSTLLRMVAGLERITSGEIWIGGRVVNELEPADRdiamv 81
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 -QNLGVCPqHnvlfdmLTVEEHIWFYARLKGLSEKHVKAEMEQMALDVGLPPSkLKSKTSQLSGGmQRKlSVAL--AFVG 1049
Cdd:PRK11650 82 fQNYALYP-H------MSVRENMAYGLKIRGMPKAEIEERVAEAARILELEPL-LDRKPRELSGG-QRQ-RVAMgrAIVR 151
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 1050 GSKVVILDEPTAGVDPYSR---RgiWELL-LKYRQGRTIILSTHHMDEADILGDRIAIISHG 1107
Cdd:PRK11650 152 EPAVFLFDEPLSNLDAKLRvqmR--LEIQrLHRRLKTTSLYVTHDQVEAMTLADRVVVMNGG 211
|
|
| PRK10789 |
PRK10789 |
SmdA family multidrug ABC transporter permease/ATP-binding protein; |
897-1127 |
1.83e-06 |
|
SmdA family multidrug ABC transporter permease/ATP-binding protein;
Pssm-ID: 182732 [Multi-domain] Cd Length: 569 Bit Score: 53.18 E-value: 1.83e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 897 LGVSIQNLVkvYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayILGKDI---RSEMSSIRQ 973
Cdd:PRK10789 314 LDVNIRQFT--YPQTDHPALENVNFTLKPGQMLGICGPTGSGKSTLLSLIQRHFDVSEGD--IRFHDIpltKLQLDSWRS 389
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 974 NLGVCPQHNVLFDMlTV------------EEHIWFYARLKGLSEKHVKaemeqmaldvgLPP---SKLKSKTSQLSGGMQ 1038
Cdd:PRK10789 390 RLAVVSQTPFLFSD-TVannialgrpdatQQEIEHVARLASVHDDILR-----------LPQgydTEVGERGVMLSGGQK 457
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1039 RKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMdEADILGDRIAIISHGKLCCVGSSLFL 1118
Cdd:PRK10789 458 QRISIARALLLNAEILILDDALSAVDGRTEHQILHNLRQWGEGRTVIISAHRL-SALTEASEILVMQHGHIAQRGNHDQL 536
|
....*....
gi 568925948 1119 KNQlgTGYY 1127
Cdd:PRK10789 537 AQQ--SGWY 543
|
|
| ArtP |
COG4161 |
ABC-type arginine transport system, ATPase component [Amino acid transport and metabolism]; |
1914-2123 |
1.87e-06 |
|
ABC-type arginine transport system, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 443326 [Multi-domain] Cd Length: 242 Bit Score: 51.55 E-value: 1.87e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLT-GDTPVT-----RGDAF-LNKNSILSNIHEV 1986
Cdd:COG4161 3 IQLKNINCFY--GSHQALFDINLECPSGETLVLLGPSGAGKSSLLRVLNlLETPDSgqlniAGHQFdFSQKPSEKAIRLL 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 HQNMGYCPQFDAITELLTGREH-VEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGP 2065
Cdd:COG4161 81 RQKVGMVFQQYNLWPHLTVMENlIEAPCKVLGLSKEQAREKAMKLLARLRLTDKADRFPLHLSGGQQQRVAIARALMMEP 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|..
gi 568925948 2066 PVVFLDEPTTGMDPKarrfLWNCALSIVKE----GRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:COG4161 161 QVLLFDEPTAALDPE----ITAQVVEIIRElsqtGITQVIVTHEVEFARKVASQVVYMEKGR 218
|
|
| PRK10253 |
PRK10253 |
iron-enterobactin ABC transporter ATP-binding protein; |
1927-2078 |
2.09e-06 |
|
iron-enterobactin ABC transporter ATP-binding protein;
Pssm-ID: 182336 [Multi-domain] Cd Length: 265 Bit Score: 51.53 E-value: 2.09e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1927 RKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNI-HEVHQNMGYCPQfDAIT----- 2000
Cdd:PRK10253 19 KYTVAENLTVEIPDGHFTAIIGPNGCGKSTLLRTLSRLMTPAHGHVWLDGEHIQHYAsKEVARRIGLLAQ-NATTpgdit 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 --ELLT-GR-EHVEFFALLRGVPEKEVGKvgewAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTG 2076
Cdd:PRK10253 98 vqELVArGRyPHQPLFTRWRKEDEEAVTK----AMQATGITHLADQSVDTLSGGQRQRAWIAMVLAQETAIMLLDEPTTW 173
|
..
gi 568925948 2077 MD 2078
Cdd:PRK10253 174 LD 175
|
|
| PRK13409 |
PRK13409 |
ribosome biogenesis/translation initiation ATPase RLI; |
1937-2104 |
2.36e-06 |
|
ribosome biogenesis/translation initiation ATPase RLI;
Pssm-ID: 184037 [Multi-domain] Cd Length: 590 Bit Score: 52.89 E-value: 2.36e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1937 GIP---PGECFGLLGVNGAGKSTTFKMLTG---------DTPVT--------RGDAFLNKNSILSN--IHEVHQnmgycP 1994
Cdd:PRK13409 92 GLPipkEGKVTGILGPNGIGKTTAVKILSGelipnlgdyEEEPSwdevlkrfRGTELQNYFKKLYNgeIKVVHK-----P 166
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1995 QF-DAITELLTG--REhveffaLLRGVPEKevGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLD 2071
Cdd:PRK13409 167 QYvDLIPKVFKGkvRE------LLKKVDER--GKLDE-VVERLGLENILDRDISELSGGELQRVAIAAALLRDADFYFFD 237
|
170 180 190
....*....|....*....|....*....|....*
gi 568925948 2072 EPTTGMDPKARrflWNCALSI--VKEGRSVVLTSH 2104
Cdd:PRK13409 238 EPTSYLDIRQR---LNVARLIreLAEGKYVLVVEH 269
|
|
| PRK11147 |
PRK11147 |
ABC transporter ATPase component; Reviewed |
933-1109 |
3.02e-06 |
|
ABC transporter ATPase component; Reviewed
Pssm-ID: 236861 [Multi-domain] Cd Length: 635 Bit Score: 52.65 E-value: 3.02e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 933 GHNGAGKTTTMSILTGLFPPTSGTaYILGKDIRseMSSIRQNlgvcPQHNV---LFDMLT-----VEEHIWFYARLKGL- 1003
Cdd:PRK11147 36 GRNGAGKSTLMKILNGEVLLDDGR-IIYEQDLI--VARLQQD----PPRNVegtVYDFVAegieeQAEYLKRYHDISHLv 108
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1004 ----SEKHVKaEME--QMALD-----------------VGLPPSKlksKTSQLSGGMQRKLSVALAFVGGSKVVILDEPT 1060
Cdd:PRK11147 109 etdpSEKNLN-ELAklQEQLDhhnlwqlenrinevlaqLGLDPDA---ALSSLSGGWLRKAALGRALVSNPDVLLLDEPT 184
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1061 AGVDpysrrgI----W-ELLLKYRQGrTIILSTHhmDEADI--LGDRIAIISHGKL 1109
Cdd:PRK11147 185 NHLD------IetieWlEGFLKTFQG-SIIFISH--DRSFIrnMATRIVDLDRGKL 231
|
|
| PLN03130 |
PLN03130 |
ABC transporter C family member; Provisional |
899-1064 |
3.13e-06 |
|
ABC transporter C family member; Provisional
Pssm-ID: 215595 [Multi-domain] Cd Length: 1622 Bit Score: 52.82 E-value: 3.13e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRS-EMSSIRQNLGV 977
Cdd:PLN03130 1238 IKFEDVVLRYRPELPPVLHGLSFEISPSEKVGIVGRTGAGKSSMLNALFRIVELERGRILIDGCDISKfGLMDLRKVLGI 1317
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLF------DMLTVEEH----IWfyarlKGLSEKHVKAEMEQMALdvGLPpSKLKSKTSQLSGGMQRKLSVALAF 1047
Cdd:PLN03130 1318 IPQAPVLFsgtvrfNLDPFNEHndadLW-----ESLERAHLKDVIRRNSL--GLD-AEVSEAGENFSVGQRQLLSLARAL 1389
|
170
....*....|....*..
gi 568925948 1048 VGGSKVVILDEPTAGVD 1064
Cdd:PLN03130 1390 LRRSKILVLDEATAAVD 1406
|
|
| PRK14247 |
PRK14247 |
phosphate ABC transporter ATP-binding protein; Provisional |
1931-2104 |
3.31e-06 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 172735 [Multi-domain] Cd Length: 250 Bit Score: 50.68 E-value: 3.31e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1931 VDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-----DTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQFDAITELLT 2004
Cdd:PRK14247 19 LDGVNLEIPDNTITALMGPSGSGKSTLLRVFNRlielyPEARVSGEVYLDGQDIFKmDVIELRRRVQMVFQIPNPIPNLS 98
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2005 GREHVEF-FALLRGVPEK-EVGKVGEWAIRKLGL---VKYG-EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK14247 99 IFENVALgLKLNRLVKSKkELQERVRWALEKAQLwdeVKDRlDAPAGKLSGGQQQRLCIARALAFQPEVLLADEPTANLD 178
|
170 180
....*....|....*....|....*.
gi 568925948 2079 PKARRFLWNCALSIVKEgRSVVLTSH 2104
Cdd:PRK14247 179 PENTAKIESLFLELKKD-MTIVLVTH 203
|
|
| PRK15064 |
PRK15064 |
ABC transporter ATP-binding protein; Provisional |
1914-2115 |
4.28e-06 |
|
ABC transporter ATP-binding protein; Provisional
Pssm-ID: 237894 [Multi-domain] Cd Length: 530 Bit Score: 51.82 E-value: 4.28e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1914 LEIKELTKIYrrKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSilsnihevhqNMGYC 1993
Cdd:PRK15064 320 LEVENLTKGF--DNGPLFKNLNLLLEAGERLAIIGENGVGKTTLLRTLVGELEPDSGTVKWSENA----------NIGYY 387
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 PQfdaitelltgrEHVEFFA----LLRGVPEKEVGKVGEWAIRK-LGLVKYGE----KYASNYSGGNKRKLSTAMALIGG 2064
Cdd:PRK15064 388 AQ-----------DHAYDFEndltLFDWMSQWRQEGDDEQAVRGtLGRLLFSQddikKSVKVLSGGEKGRMLFGKLMMQK 456
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2065 PPVVFLDEPTTGMDPKARRFLwNCALSiVKEGrSVVLTSHSMEECEALCTR 2115
Cdd:PRK15064 457 PNVLVMDEPTNHMDMESIESL-NMALE-KYEG-TLIFVSHDREFVSSLATR 504
|
|
| PRK15112 |
PRK15112 |
peptide ABC transporter ATP-binding protein SapF; |
1913-2104 |
4.50e-06 |
|
peptide ABC transporter ATP-binding protein SapF;
Pssm-ID: 185067 [Multi-domain] Cd Length: 267 Bit Score: 50.56 E-value: 4.50e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYR-------RKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDaflnknsILSNIHE 1985
Cdd:PRK15112 4 LLEVRNLSKTFRyrtgwfrRQTVEAVKPLSFTLREGQTLAIIGENGSGKSTLAKMLAGMIEPTSGE-------LLIDDHP 76
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VH-QNMGYCPQ-----FDAITELLTGREHV----EFFALLR---GVPEKEvgKVGEWAIRKLGLVK-YGEKYASNYSGGN 2051
Cdd:PRK15112 77 LHfGDYSYRSQrirmiFQDPSTSLNPRQRIsqilDFPLRLNtdlEPEQRE--KQIIETLRQVGLLPdHASYYPHMLAPGQ 154
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2052 KRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIV-KEGRS-VVLTSH 2104
Cdd:PRK15112 155 KQRLGLARALILRPKVIIADEALASLDMSMRSQLINLMLELQeKQGISyIYVTQH 209
|
|
| PRK15134 |
PRK15134 |
microcin C ABC transporter ATP-binding protein YejF; Provisional |
911-1080 |
4.65e-06 |
|
microcin C ABC transporter ATP-binding protein YejF; Provisional
Pssm-ID: 237917 [Multi-domain] Cd Length: 529 Bit Score: 51.63 E-value: 4.65e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 911 GMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPpTSGTAYILGKDI----RSEMSSIRQNLGVCPQ--HNVL 984
Cdd:PRK15134 297 DHNVVVKNISFTLRPGETLGLVGESGSGKSTTGLALLRLIN-SQGEIWFDGQPLhnlnRRQLLPVRHRIQVVFQdpNSSL 375
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 985 FDMLTVEEHIWFYARL--KGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAG 1062
Cdd:PRK15134 376 NPRLNVLQIIEEGLRVhqPTLSAAQREQQVIAVMEEVGLDPETRHRYPAEFSGGQRQRIAIARALILKPSLIILDEPTSS 455
|
170
....*....|....*...
gi 568925948 1063 VDPYSRRGIWELLLKYRQ 1080
Cdd:PRK15134 456 LDKTVQAQILALLKSLQQ 473
|
|
| PRK10938 |
PRK10938 |
putative molybdenum transport ATP-binding protein ModF; Provisional |
899-1093 |
4.69e-06 |
|
putative molybdenum transport ATP-binding protein ModF; Provisional
Pssm-ID: 182852 [Multi-domain] Cd Length: 490 Bit Score: 51.55 E-value: 4.69e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYIL-GK---------DIRSEM 968
Cdd:PRK10938 261 IVLNNGVVSYND--RPILHNLSWQVNPGEHWQIVGPNGAGKSTLLSLITGDHPQGYSNDLTLfGRrrgsgetiwDIKKHI 338
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 ----SSIRQNLGV-CPQHNVL----FDmltveeHIWFYarlKGLSEKHVKAEMEQMALdVGLPPSKLKSKTSQLSGGMQR 1039
Cdd:PRK10938 339 gyvsSSLHLDYRVsTSVRNVIlsgfFD------SIGIY---QAVSDRQQKLAQQWLDI-LGIDKRTADAPFHSLSWGQQR 408
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*..
gi 568925948 1040 KLSVALAFVGGSKVVILDEPTAGVDPYSR---RGIWELLLkyRQGRTIILSTHHMDE 1093
Cdd:PRK10938 409 LALIVRALVKHPTLLILDEPLQGLDPLNRqlvRRFVDVLI--SEGETQLLFVSHHAE 463
|
|
| ycf16 |
CHL00131 |
sulfate ABC transporter protein; Validated |
1913-2104 |
4.97e-06 |
|
sulfate ABC transporter protein; Validated
Pssm-ID: 214372 [Multi-domain] Cd Length: 252 Bit Score: 50.41 E-value: 4.97e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELtkiyrrkrKPAVDRICI------GIPPGECFGLLGVNGAGKSTTFKMLTG--DTPVTRGDAFLNKNSILSNIH 1984
Cdd:CHL00131 7 ILEIKNL--------HASVNENEIlkglnlSINKGEIHAIMGPNGSGKSTLSKVIAGhpAYKILEGDILFKGESILDLEP 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1985 EVHQNMGYCPQFDAITElLTGREHVEFFAL-------LRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNY-----SGGNK 2052
Cdd:CHL00131 79 EERAHLGIFLAFQYPIE-IPGVSNADFLRLaynskrkFQGLPELDPLEFLEIINEKLKLVGMDPSFLSRNvnegfSGGEK 157
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2053 RK---LStaMALIgGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:CHL00131 158 KRneiLQ--MALL-DSELAILDETDSGLDIDALKIIAEGINKLMTSENSIILITH 209
|
|
| ABC_ABC_ChvD |
TIGR03719 |
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of ... |
1913-2078 |
6.75e-06 |
|
ATP-binding cassette protein, ChvD family; Members of this protein family have two copies of the ABC transporter ATP-binding cassette, but are found outside the common ABC transporter operon structure that features integral membrane permease proteins and substrate-binding proteins encoded next to the ATP-binding cassette (ABC domain) protein. The member protein ChvD from Agrobacterium tumefaciens was identified as both a candidate to interact with VirB8, based on yeast two-hybrid analysis, and as an apparent regulator of VirG. The general function of this protein family is unknown.
Pssm-ID: 274744 [Multi-domain] Cd Length: 552 Bit Score: 51.09 E-value: 6.75e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPVTrGDAFLNKNSilsnihevhqNMG 1991
Cdd:TIGR03719 4 IYTMNRVSKVVPPKKE-ILKDISLSFFPGAKIGVLGLNGAGKSTLLRIMAGvDKDFN-GEARPQPGI----------KVG 71
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1992 YCPQFDAITELLTGREHVE--------------------------FFALLR--GVPEKEVGKVGEWAI-RKLGLVKY--- 2039
Cdd:TIGR03719 72 YLPQEPQLDPTKTVRENVEegvaeikdaldrfneisakyaepdadFDKLAAeqAELQEIIDAADAWDLdSQLEIAMDalr 151
|
170 180 190 200
....*....|....*....|....*....|....*....|..
gi 568925948 2040 ---GEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:TIGR03719 152 cppWDADVTKLSGGERRRVALCRLLLSKPDMLLLDEPTNHLD 193
|
|
| thiQ |
PRK10771 |
thiamine ABC transporter ATP-binding protein ThiQ; |
1938-2133 |
6.88e-06 |
|
thiamine ABC transporter ATP-binding protein ThiQ;
Pssm-ID: 182716 [Multi-domain] Cd Length: 232 Bit Score: 49.58 E-value: 6.88e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKN------------SIL---SNI--H-EVHQN--MGYCPQFD 1997
Cdd:PRK10771 22 VERGERVAILGPSGAGKSTLLNLIAGFLTPASGSLTLNGQdhtttppsrrpvSMLfqeNNLfsHlTVAQNigLGLNPGLK 101
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1998 aitelLTGREHveffallrgvpekevGKVGEWAiRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGM 2077
Cdd:PRK10771 102 -----LNAAQR---------------EKLHAIA-RQMGIEDLLARLPGQLSGGQRQRVALARCLVREQPILLLDEPFSAL 160
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|.
gi 568925948 2078 DPKARrflwNCALSIVKE---GRSVVL--TSHSMEECEALCTRMAIMVNGRFRCLGSVQHL 2133
Cdd:PRK10771 161 DPALR----QEMLTLVSQvcqERQLTLlmVSHSLEDAARIAPRSLVVADGRIAWDGPTDEL 217
|
|
| znuC |
PRK09544 |
high-affinity zinc transporter ATPase; Reviewed |
1934-2106 |
7.31e-06 |
|
high-affinity zinc transporter ATPase; Reviewed
Pssm-ID: 181939 [Multi-domain] Cd Length: 251 Bit Score: 49.73 E-value: 7.31e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1934 ICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknsilSNIHEVHQNMGYCPQ---FDAITELLTGRehve 2010
Cdd:PRK09544 23 VSLELKPGKILTLLGPNGAGKSTLVRVVLGLVAPDEG----------VIKRNGKLRIGYVPQklyLDTTLPLTVNR---- 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2011 FFALLRGVPEKEVGKvgewAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCAL 2090
Cdd:PRK09544 89 FLRLRPGTKKEDILP----ALKRVQAGHLIDAPMQKLSGGETQRVLLARALLNRPQLLVLDEPTQGVDVNGQVALYDLID 164
|
170
....*....|....*..
gi 568925948 2091 SIVKE-GRSVVLTSHSM 2106
Cdd:PRK09544 165 QLRRElDCAVLMVSHDL 181
|
|
| PLN03130 |
PLN03130 |
ABC transporter C family member; Provisional |
926-1109 |
8.11e-06 |
|
ABC transporter C family member; Provisional
Pssm-ID: 215595 [Multi-domain] Cd Length: 1622 Bit Score: 51.66 E-value: 8.11e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYIlgkdirsemssIRQNLGVCPQHNVLFDMlTVEEHIWFYARLKglSE 1005
Cdd:PLN03130 643 GSLVAIVGSTGEGKTSLISAMLGELPPRSDASVV-----------IRGTVAYVPQVSWIFNA-TVRDNILFGSPFD--PE 708
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1006 KHVKA-EMEQMALDVGLPP----SKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYR- 1079
Cdd:PLN03130 709 RYERAiDVTALQHDLDLLPggdlTEIGERGVNISGGQKQRVSMARAVYSNSDVYIFDDPLSALDAHVGRQVFDKCIKDEl 788
|
170 180 190
....*....|....*....|....*....|...
gi 568925948 1080 QGRTIILST---HHMDEAdilgDRIAIISHGKL 1109
Cdd:PLN03130 789 RGKTRVLVTnqlHFLSQV----DRIILVHEGMI 817
|
|
| PRK10636 |
PRK10636 |
putative ABC transporter ATP-binding protein; Provisional |
1931-2104 |
9.40e-06 |
|
putative ABC transporter ATP-binding protein; Provisional
Pssm-ID: 236729 [Multi-domain] Cd Length: 638 Bit Score: 50.94 E-value: 9.40e-06
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1931 VDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILsnihevhqnmGYCPQFDaiTELLTGRE--- 2007
Cdd:PRK10636 328 LDSIKLNLVPGSRIGLLGRNGAGKSTLIKLLAGELAPVSGEIGLAKGIKL----------GYFAQHQ--LEFLRADEspl 395
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2008 -HveffaLLRGVPekevgKVGEWAIRK-LGLVKY-GEKYASN---YSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKA 2081
Cdd:PRK10636 396 qH-----LARLAP-----QELEQKLRDyLGGFGFqGDKVTEEtrrFSGGEKARLVLALIVWQRPNLLLLDEPTNHLDLDM 465
|
170 180
....*....|....*....|...
gi 568925948 2082 RRFLWNcALsIVKEGrSVVLTSH 2104
Cdd:PRK10636 466 RQALTE-AL-IDFEG-ALVVVSH 485
|
|
| PRK10982 |
PRK10982 |
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional |
919-1109 |
1.07e-05 |
|
galactose/methyl galaxtoside transporter ATP-binding protein; Provisional
Pssm-ID: 182880 [Multi-domain] Cd Length: 491 Bit Score: 50.50 E-value: 1.07e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 919 LALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI--RSEMSSI----------RQNLGVCPQHNVLFD 986
Cdd:PRK10982 267 VSFDLHKGEILGIAGLVGAKRTDIVETLFGIREKSAGTITLHGKKInnHNANEAInhgfalvteeRRSTGIYAYLDIGFN 346
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 MLtVEEHIWFYARLKGLSEKHVKAEMEQM--ALDVGLPPSKlkSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVD 1064
Cdd:PRK10982 347 SL-ISNIRNYKNKVGLLDNSRMKSDTQWVidSMRVKTPGHR--TQIGSLSGGNQQKVIIGRWLLTQPEILMLDEPTRGID 423
|
170 180 190 200
....*....|....*....|....*....|....*....|....*...
gi 568925948 1065 PYSRRGIWELLLKY-RQGRTIILSTHHMDEadILG--DRIAIISHGKL 1109
Cdd:PRK10982 424 VGAKFEIYQLIAELaKKDKGIIIISSEMPE--LLGitDRILVMSNGLV 469
|
|
| AppF |
COG4608 |
ABC-type oligopeptide transport system, ATPase component [Amino acid transport and metabolism]; ... |
1907-2078 |
1.14e-05 |
|
ABC-type oligopeptide transport system, ATPase component [Amino acid transport and metabolism];
Pssm-ID: 443658 [Multi-domain] Cd Length: 329 Bit Score: 49.73 E-value: 1.14e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1907 GGGQNDILEIKELTKIY-------RRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKN 1977
Cdd:COG4608 1 AAMAEPLLEVRDLKKHFpvrgglfGRTVGVvkAVDGVSFDIRRGETLGLVGESGCGKSTLGRLLLRLEEPTSGEILFDGQ 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1978 SILS-----------NIHEVHQNmgycPQ---------FDAITELLtgREHveffallRGVPEKEV-GKVGEwAIRKLGL 2036
Cdd:COG4608 81 DITGlsgrelrplrrRMQMVFQD----PYaslnprmtvGDIIAEPL--RIH-------GLASKAERrERVAE-LLELVGL 146
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 2037 -VKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:COG4608 147 rPEHADRYPHEFSGGQRQRIGIARALALNPKLIVCDEPVSALD 189
|
|
| AAA_21 |
pfam13304 |
AAA domain, putative AbiEii toxin, Type IV TA system; Several members are annotated as being ... |
1031-1089 |
1.26e-05 |
|
AAA domain, putative AbiEii toxin, Type IV TA system; Several members are annotated as being of the abortive phage resistance system, in which case the family would be acting as the toxin for a type IV toxin-antitoxin resistance system.
Pssm-ID: 433102 [Multi-domain] Cd Length: 303 Bit Score: 49.70 E-value: 1.26e-05
10 20 30 40 50 60
....*....|....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1031 SQLSGGMQRKLSVALAF---VGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRT-IILSTH 1089
Cdd:pfam13304 235 FELSDGTKRLLALLAALlsaLPKGGLLLIDEPESGLHPKLLRRLLELLKELSRNGAqLILTTH 297
|
|
| PRK10535 |
PRK10535 |
macrolide ABC transporter ATP-binding protein/permease MacB; |
1913-2104 |
1.29e-05 |
|
macrolide ABC transporter ATP-binding protein/permease MacB;
Pssm-ID: 182528 [Multi-domain] Cd Length: 648 Bit Score: 50.49 E-value: 1.29e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKRKP--AVDRICIGIPPGECFGLLGVNGAGKSTTFKML-------TGDTPVTRGDAFLNKNSILSNI 1983
Cdd:PRK10535 4 LLELKDIRRSYPSGEEQveVLKGISLDIYAGEMVAIVGASGSGKSTLMNILgcldkptSGTYRVAGQDVATLDADALAQL 83
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1984 HEVHqnMGYCPQFDAITELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIG 2063
Cdd:PRK10535 84 RREH--FGFIFQRYHLLSHLTAAQNVEVPAVYAGLERKQRLLRAQELLQRLGLEDRVEYQPSQLSGGQQQRVSIARALMN 161
|
170 180 190 200
....*....|....*....|....*....|....*....|.
gi 568925948 2064 GPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:PRK10535 162 GGQVILADEPTGALDSHSGEEVMAILHQLRDRGHTVIIVTH 202
|
|
| PTZ00265 |
PTZ00265 |
multidrug resistance protein (mdr1); Provisional |
872-1091 |
2.39e-05 |
|
multidrug resistance protein (mdr1); Provisional
Pssm-ID: 240339 [Multi-domain] Cd Length: 1466 Bit Score: 50.03 E-value: 2.39e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 872 EKSHPGSSQKGVSEIcmeeEPTHLRLGVSIQNLVKVYRDgmkvavdgLALNFYEGQITSFLGHNGAGKTTTMSILTGLFP 951
Cdd:PTZ00265 369 ENNDDGKKLKDIKKI----QFKNVRFHYDTRKDVEIYKD--------LNFTLTEGKTYAFVGESGCGKSTILKLIERLYD 436
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 952 PTSGTAYILG----KDIrsEMSSIRQNLGVCPQHNVLFDML---TVEEHIWFYARLKGLSEK------------------ 1006
Cdd:PTZ00265 437 PTEGDIIINDshnlKDI--NLKWWRSKIGVVSQDPLLFSNSiknNIKYSLYSLKDLEALSNYynedgndsqenknkrnsc 514
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1007 ---------------------HVKAEMEQM----ALDV-----------GLP---PSKLKSKTSQLSGGMQRKLSVALAF 1047
Cdd:PTZ00265 515 rakcagdlndmsnttdsneliEMRKNYQTIkdseVVDVskkvlihdfvsALPdkyETLVGSNASKLSGGQKQRISIARAI 594
|
250 260 270 280
....*....|....*....|....*....|....*....|....*.
gi 568925948 1048 VGGSKVVILDEPTAGVDPYSRRGIWELL--LKYRQGRTIILSTHHM 1091
Cdd:PTZ00265 595 IRNPKILILDEATSSLDNKSEYLVQKTInnLKGNENRITIIIAHRL 640
|
|
| PRK10636 |
PRK10636 |
putative ABC transporter ATP-binding protein; Provisional |
909-1109 |
2.82e-05 |
|
putative ABC transporter ATP-binding protein; Provisional
Pssm-ID: 236729 [Multi-domain] Cd Length: 638 Bit Score: 49.40 E-value: 2.82e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 909 RDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTG---------LFP---------------PTSGTAYILGKDi 964
Cdd:PRK10636 10 RRGVRVLLDNATATINPGQKVGLVGKNGCGKSTLLALLKNeisadggsyTFPgnwqlawvnqetpalPQPALEYVIDGD- 88
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 965 rSEMSSIRQNLGVCPQHNVLFDMLTVeehiwfYARLKGLSEKHVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVA 1044
Cdd:PRK10636 89 -REYRQLEAQLHDANERNDGHAIATI------HGKLDAIDAWTIRSRAASLLHGLGFSNEQLERPVSDFSGGWRMRLNLA 161
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1045 LAFVGGSKVVILDEPTAGVDPYSRrgIW-ELLLKYRQGrTIILSTHHMDEADILGDRIAIISHGKL 1109
Cdd:PRK10636 162 QALICRSDLLLLDEPTNHLDLDAV--IWlEKWLKSYQG-TLILISHDRDFLDPIVDKIIHIEQQSL 224
|
|
| PTZ00243 |
PTZ00243 |
ABC transporter; Provisional |
913-1115 |
3.04e-05 |
|
ABC transporter; Provisional
Pssm-ID: 240327 [Multi-domain] Cd Length: 1560 Bit Score: 49.39 E-value: 3.04e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 913 KVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYilgkdirSEMSsirqnLGVCPQHNVLFDMlTVEE 992
Cdd:PTZ00243 673 KVLLRDVSVSVPRGKLTVVLGATGSGKSTLLQSLLSQFEISEGRVW-------AERS-----IAYVPQQAWIMNA-TVRG 739
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 993 HIWFY-----ARLkglsekHVKAEMEQMALDVGLPPSKLKS----KTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGV 1063
Cdd:PTZ00243 740 NILFFdeedaARL------ADAVRVSQLEADLAQLGGGLETeigeKGVNLSGGQKARVSLARAVYANRDVYLLDDPLSAL 813
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|...
gi 568925948 1064 DPY-SRRGIWELLLKYRQGRTIILSTHHMDEADiLGDRIAIISHGKLCCVGSS 1115
Cdd:PTZ00243 814 DAHvGERVVEECFLGALAGKTRVLATHQVHVVP-RADYVVALGDGRVEFSGSS 865
|
|
| PTZ00243 |
PTZ00243 |
ABC transporter; Provisional |
908-1114 |
3.25e-05 |
|
ABC transporter; Provisional
Pssm-ID: 240327 [Multi-domain] Cd Length: 1560 Bit Score: 49.39 E-value: 3.25e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 908 YRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDIRSE-MSSIRQNLGVCPQHNVLFD 986
Cdd:PTZ00243 1318 YREGLPLVLRGVSFRIAPREKVGIVGRTGSGKSTLLLTFMRMVEVCGGEIRVNGREIGAYgLRELRRQFSMIPQDPVLFD 1397
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 987 ---MLTVE-------EHIWFYARLKGLSEkHVKAEMEqmALDvglppSKLKSKTSQLSGGMQRKLSVALAFVG-GSKVVI 1055
Cdd:PTZ00243 1398 gtvRQNVDpfleassAEVWAALELVGLRE-RVASESE--GID-----SRVLEGGSNYSVGQRQLMCMARALLKkGSGFIL 1469
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 1056 LDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILgDRIAIISHGKLCCVGS 1114
Cdd:PTZ00243 1470 MDEATANIDPALDRQIQATVMSAFSAYTVITIAHRLHTVAQY-DKIIVMDHGAVAEMGS 1527
|
|
| Rli1 |
COG1245 |
Translation initiation factor RLI1, contains Fe-S and AAA+ ATPase domains [Translation, ... |
888-1104 |
4.04e-05 |
|
Translation initiation factor RLI1, contains Fe-S and AAA+ ATPase domains [Translation, ribosomal structure and biogenesis];
Pssm-ID: 440858 [Multi-domain] Cd Length: 592 Bit Score: 48.63 E-value: 4.04e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 888 MEEEPTHlRLGvsiQNLVKVYrdGMKVAVdglalnfyEGQITSFLGHNGAGKTTTMSILTGLFPPTSG-------TAYIL 960
Cdd:COG1245 75 LEEDPVH-RYG---ENGFRLY--GLPVPK--------KGKVTGILGPNGIGKSTALKILSGELKPNLGdydeepsWDEVL 140
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 961 ----GKDIRSEMSSIRQN-LGVC--PQHnvlFDML------TVEEhiwfyaRLKGLSEKHVKAE-MEQMALDvglppSKL 1026
Cdd:COG1245 141 krfrGTELQDYFKKLANGeIKVAhkPQY---VDLIpkvfkgTVRE------LLEKVDERGKLDElAEKLGLE-----NIL 206
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1027 KSKTSQLSGG-MQRkLSVALAFVGGSKVVILDEPTAGVDPYSR----RGIWELLlkyRQGRTIILSTHHMDEADILGDRI 1101
Cdd:COG1245 207 DRDISELSGGeLQR-VAIAAALLRDADFYFFDEPSSYLDIYQRlnvaRLIRELA---EEGKYVLVVEHDLAILDYLADYV 282
|
...
gi 568925948 1102 AII 1104
Cdd:COG1245 283 HIL 285
|
|
| PRK11819 |
PRK11819 |
putative ABC transporter ATP-binding protein; Reviewed |
1890-2078 |
4.37e-05 |
|
putative ABC transporter ATP-binding protein; Reviewed
Pssm-ID: 236992 [Multi-domain] Cd Length: 556 Bit Score: 48.58 E-value: 4.37e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1890 LNDEDEDVRRERQRILDGGGQ---NDILEIKELTKIYRrkrkpavDRICI-----GIPPGECFGLLGVNGAGKSTTFKML 1961
Cdd:PRK11819 298 LLSEEYQKRNETNEIFIPPGPrlgDKVIEAENLSKSFG-------DRLLIddlsfSLPPGGIVGIIGPNGAGKSTLFKMI 370
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1962 TG-DTP----VTRGDAflnknsilsniheVHqnMGYCPQFdaitelltgREHVEffallrgvPEKEVGKV--GEWAIRKL 2034
Cdd:PRK11819 371 TGqEQPdsgtIKIGET-------------VK--LAYVDQS---------RDALD--------PNKTVWEEisGGLDIIKV 418
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2035 GLVK-----Y-------G---EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK11819 419 GNREipsraYvgrfnfkGgdqQKKVGVLSGGERNRLHLAKTLKQGGNVLLLDEPTNDLD 477
|
|
| MK0520 |
COG2401 |
ABC-type ATPase fused to a predicted acetyltransferase domain [General function prediction ... |
1923-2104 |
4.82e-05 |
|
ABC-type ATPase fused to a predicted acetyltransferase domain [General function prediction only];
Pssm-ID: 441957 [Multi-domain] Cd Length: 222 Bit Score: 46.88 E-value: 4.82e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1923 YRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG---DTPVT-----RGDAFLNKNSILSNIhevhqnmGYCP 1994
Cdd:COG2401 38 LRVVERYVLRDLNLEIEPGEIVLIVGASGSGKSTLLRLLAGalkGTPVAgcvdvPDNQFGREASLIDAI-------GRKG 110
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1995 QFDAITELLTGREHVEFFALLRgvpekevgkvgewairklglvKYGEkyasnYSGGNKRKLSTAMALIGGPPVVFLDEPT 2074
Cdd:COG2401 111 DFKDAVELLNAVGLSDAVLWLR---------------------RFKE-----LSTGQKFRFRLALLLAERPKLLVIDEFC 164
|
170 180 190
....*....|....*....|....*....|.
gi 568925948 2075 TGMDPK-ARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:COG2401 165 SHLDRQtAKRVARNLQKLARRAGITLVVATH 195
|
|
| GguA |
NF040905 |
sugar ABC transporter ATP-binding protein; |
2044-2124 |
5.25e-05 |
|
sugar ABC transporter ATP-binding protein;
Pssm-ID: 468840 [Multi-domain] Cd Length: 500 Bit Score: 48.25 E-value: 5.25e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2044 ASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHSMEECEALCTRMAIMVNGR 2123
Cdd:NF040905 402 VGNLSGGNQQKVVLSKWLFTDPDVLILDEPTRGIDVGAKYEIYTIINELAAEGKGVIVISSELPELLGMCDRIYVMNEGR 481
|
.
gi 568925948 2124 F 2124
Cdd:NF040905 482 I 482
|
|
| ABC_UvrA_II |
cd03271 |
ATP-binding cassette domain II of the excision repair protein UvrA; Nucleotide excision repair ... |
981-1092 |
9.94e-05 |
|
ATP-binding cassette domain II of the excision repair protein UvrA; Nucleotide excision repair in eubacteria is a process that repairs DNA damage by the removal of a 12-13-mer oligonucleotide containing the lesion. Recognition and cleavage of the damaged DNA is a multistep ATP-dependent reaction that requires the UvrA, UvrB, and UvrC proteins. Both UvrA and UvrB are ATPases, with UvrA having two ATP binding sites, which have the characteristic signature of the family of ABC proteins and UvrB having one ATP binding site that is structurally related to that of helicases.
Pssm-ID: 213238 [Multi-domain] Cd Length: 261 Bit Score: 46.45 E-value: 9.94e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLfDMlTVEEHIWFYARLKGLSEKHvkaemeQMALDVGLPPSKLKSKTSQLSGG-MQR-KLSVALAF-VGGSKVVILD 1057
Cdd:cd03271 126 ADVL-DM-TVEEALEFFENIPKIARKL------QTLCDVGLGYIKLGQPATTLSGGeAQRiKLAKELSKrSTGKTLYILD 197
|
90 100 110
....*....|....*....|....*....|....*.
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMD 1092
Cdd:cd03271 198 EPTTGLHFHDVKKLLEVLQRLVdKGNTVVVIEHNLD 233
|
|
| GguA |
NF040905 |
sugar ABC transporter ATP-binding protein; |
916-1109 |
1.06e-04 |
|
sugar ABC transporter ATP-binding protein;
Pssm-ID: 468840 [Multi-domain] Cd Length: 500 Bit Score: 47.48 E-value: 1.06e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLALNFYEGQITSFLGHNGAGKT-TTMSILT---GLFppTSGTAYILGKDIRseMSSI--------------RQNLGV 977
Cdd:NF040905 276 VDDVSLNVRRGEIVGIAGLMGAGRTeLAMSVFGrsyGRN--ISGTVFKDGKEVD--VSTVsdaidaglayvtedRKGYGL 351
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 cpqhnVLFDmlTVEEHIWFyARLKGLSEKHVKAEMEQMALDVGLPpSKLKSKTS-------QLSGGMQRKlsVALA---F 1047
Cdd:NF040905 352 -----NLID--DIKRNITL-ANLGKVSRRGVIDENEEIKVAEEYR-KKMNIKTPsvfqkvgNLSGGNQQK--VVLSkwlF 420
|
170 180 190 200 210 220
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 1048 VGgSKVVILDEPTAGVDPYSRRGIWELLLKY-RQGRTIILSTHHMDEadILG--DRIAIISHGKL 1109
Cdd:NF040905 421 TD-PDVLILDEPTRGIDVGAKYEIYTIINELaAEGKGVIVISSELPE--LLGmcDRIYVMNEGRI 482
|
|
| AAA |
smart00382 |
ATPases associated with a variety of cellular activities; AAA - ATPases associated with a ... |
926-1104 |
1.40e-04 |
|
ATPases associated with a variety of cellular activities; AAA - ATPases associated with a variety of cellular activities. This profile/alignment only detects a fraction of this vast family. The poorly conserved N-terminal helix is missing from the alignment.
Pssm-ID: 214640 [Multi-domain] Cd Length: 148 Bit Score: 44.29 E-value: 1.40e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 926 GQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYIlgkdirsemssirqnlgvcpqhnvlfdmltveehiwfyarlkglse 1005
Cdd:smart00382 2 GEVILIVGPPGSGKTTLARALARELGPPGGGVIY---------------------------------------------- 35
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1006 khVKAEMEQMALDVGLPPSKLKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELL-------LKY 1078
Cdd:smart00382 36 --IDGEDILEEVLDQLLLIIVGGKKASGSGELRLRLALALARKLKPDVLILDEITSLLDAEQEALLLLLEelrllllLKS 113
|
170 180
....*....|....*....|....*.
gi 568925948 1079 RQGRTIILSTHHMdeaDILGDRIAII 1104
Cdd:smart00382 114 EKNLTVILTTNDE---KDLGPALLRR 136
|
|
| 3a01208 |
TIGR00958 |
Conjugate Transporter-2 (CT2) Family protein; [Transport and binding proteins, Other] |
1938-2144 |
1.58e-04 |
|
Conjugate Transporter-2 (CT2) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273363 [Multi-domain] Cd Length: 711 Bit Score: 47.02 E-value: 1.58e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILS-NIHEVHQNMGYCPQfdaiTELLTGREHVEFFAL-L 2015
Cdd:TIGR00958 504 LHPGEVVALVGPSGSGKSTVAALLQNLYQPTGGQVLLDGVPLVQyDHHYLHRQVALVGQ----EPVLFSGSVRENIAYgL 579
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2016 RGVPEKEVGKVGEWA-----IRKLGLVKY---GEKyASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARRFL-- 2085
Cdd:TIGR00958 580 TDTPDEEIMAAAKAAnahdfIMEFPNGYDtevGEK-GSQLSGGQKQRIAIARALVRKPRVLILDEATSALDAECEQLLqe 658
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2086 WNCAlsivkEGRSVVLTSHSMEECEAlCTRMAIMVNGRFRCLGSVQHLKNRFGDGYTIV 2144
Cdd:TIGR00958 659 SRSR-----ASRTVLLIAHRLSTVER-ADQILVLKKGSVVEMGTHKQLMEDQGCYKHLV 711
|
|
| ModC |
COG4148 |
ABC-type molybdate transport system, ATPase component ModC [Inorganic ion transport and ... |
1938-2170 |
1.74e-04 |
|
ABC-type molybdate transport system, ATPase component ModC [Inorganic ion transport and metabolism]; ABC-type molybdate transport system, ATPase component ModC is part of the Pathway/BioSystem: Molybdopterin biosynthesis
Pssm-ID: 443319 [Multi-domain] Cd Length: 358 Bit Score: 46.25 E-value: 1.74e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNkNSILSNIHE-----VHQ-NMGYCPQfDAitEL---LTGREH 2008
Cdd:COG4148 22 LPGRGVTALFGPSGSGKTTLLRAIAGLERPDSGRIRLG-GEVLQDSARgiflpPHRrRIGYVFQ-EA--RLfphLSVRGN 97
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2009 VEFfALLRGVPEKEVGKVGEwAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKARR----F 2084
Cdd:COG4148 98 LLY-GRKRAPRAERRISFDE-VVELLGIGHLLDRRPATLSGGERQRVAIGRALLSSPRLLLMDEPLAALDLARKAeilpY 175
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2085 L--WNCALSIvkegrSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRfgdgytivvriagsnPDLKPVQEFFG 2162
Cdd:COG4148 176 LerLRDELDI-----PILYVSHSLDEVARLADHVVLLEQGRVVASGPLAEVLSR---------------PDLLPLAGGEE 235
|
....*...
gi 568925948 2163 lafPGSVL 2170
Cdd:COG4148 236 ---AGSVL 240
|
|
| MRP_assoc_pro |
TIGR00957 |
multi drug resistance-associated protein (MRP); This model describes multi drug ... |
899-1064 |
2.31e-04 |
|
multi drug resistance-associated protein (MRP); This model describes multi drug resistance-associated protein (MRP) in eukaryotes. The multidrug resistance-associated protein is an integral membrane protein that causes multidrug resistance when overexpressed in mammalian cells. It belongs to ABC transporter superfamily. The protein topology and function was experimentally demonstrated by epitope tagging and immunofluorescence. Insertion of tags in the critical regions associated with drug efflux, abrogated its function. The C-terminal domain seem to highly conserved. [Transport and binding proteins, Other]
Pssm-ID: 188098 [Multi-domain] Cd Length: 1522 Bit Score: 46.86 E-value: 2.31e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTttmSILTGLF---PPTSGTAYILGKDI-RSEMSSIRQN 974
Cdd:TIGR00957 1285 VEFRNYCLRYREDLDLVLRHINVTIHGGEKVGIVGRTGAGKS---SLTLGLFrinESAEGEIIIDGLNIaKIGLHDLRFK 1361
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 975 LGVCPQHNVLF--------DMLT--VEEHIWFyarlkGLSEKHVKAEMEQMaldvglpPSKLKSKTSQ----LSGGMQRK 1040
Cdd:TIGR00957 1362 ITIIPQDPVLFsgslrmnlDPFSqySDEEVWW-----ALELAHLKTFVSAL-------PDKLDHECAEggenLSVGQRQL 1429
|
170 180
....*....|....*....|....
gi 568925948 1041 LSVALAFVGGSKVVILDEPTAGVD 1064
Cdd:TIGR00957 1430 VCLARALLRKTKILVLDEATAAVD 1453
|
|
| ABC_UvrA_I |
cd03270 |
ATP-binding cassette domain I of the excision repair protein UvrA; Nucleotide excision repair ... |
1014-1101 |
2.37e-04 |
|
ATP-binding cassette domain I of the excision repair protein UvrA; Nucleotide excision repair in eubacteria is a process that repairs DNA damage by the removal of a 12-13-mer oligonucleotide containing the lesion. Recognition and cleavage of the damaged DNA is a multistep ATP-dependent reaction that requires the UvrA, UvrB, and UvrC proteins. Both UvrA and UvrB are ATPases, with UvrA having two ATP binding sites, which have the characteristic signature of the family of ABC proteins, and UvrB having one ATP binding site that is structurally related to that of helicases.
Pssm-ID: 213237 [Multi-domain] Cd Length: 226 Bit Score: 44.94 E-value: 2.37e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1014 QMALDVGLPPSKLKSKTSQLSGG-MQRklsVALAFVGGSKVV----ILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILS 1087
Cdd:cd03270 119 GFLVDVGLGYLTLSRSAPTLSGGeAQR---IRLATQIGSGLTgvlyVLDEPSIGLHPRDNDRLIETLKRLRdLGNTVLVV 195
|
90
....*....|....*
gi 568925948 1088 THhmDEADIL-GDRI 1101
Cdd:cd03270 196 EH--DEDTIRaADHV 208
|
|
| ABCD_peroxisomal_ALDP |
cd03223 |
ATP-binding cassette domain of peroxisomal transporter, subfamily D; Peroxisomal ATP-binding ... |
899-1106 |
2.81e-04 |
|
ATP-binding cassette domain of peroxisomal transporter, subfamily D; Peroxisomal ATP-binding cassette transporter (Pat) is involved in the import of very long-chain fatty acids (VLCFA) into the peroxisome. The peroxisomal membrane forms a permeability barrier for a wide variety of metabolites required for and formed during fatty acid beta-oxidation. To communicate with the cytoplasm and mitochondria, peroxisomes need dedicated proteins to transport such hydrophilic molecules across their membranes. X-linked adrenoleukodystrophy (X-ALD) is caused by mutations in the ALD gene, which encodes ALDP (adrenoleukodystrophy protein ), a peroxisomal integral membrane protein that is a member of the ATP-binding cassette (ABC) transporter protein family. The disease is characterized by a striking and unpredictable variation in phenotypic expression. Phenotypes include the rapidly progressive childhood cerebral form (CCALD), the milder adult form, adrenomyeloneuropathy (AMN), and variants without neurologic involvement (i.e. asymptomatic).
Pssm-ID: 213190 [Multi-domain] Cd Length: 166 Bit Score: 43.68 E-value: 2.81e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGmKVAVDGLALNFYEGQ---ITsflGHNGAGKTTTMSILTGLFPPTSGTayilgkdirsemssirqnL 975
Cdd:cd03223 1 IELENLSLATPDG-RVLLKDLSFEIKPGDrllIT---GPSGTGKSSLFRALAGLWPWGSGR------------------I 58
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 976 GVCPQHNVLFdmltveehiwfyarlkglsekhvkaeMEQMALdvgLPPSKLKSK-----TSQLSGGMQRKLSVALAFVGG 1050
Cdd:cd03223 59 GMPEGEDLLF--------------------------LPQRPY---LPLGTLREQliypwDDVLSGGEQQRLAFARLLLHK 109
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....*.
gi 568925948 1051 SKVVILDEPTAGVDPYSRRGIWELLlkyRQGRTIILSTHHMDEADILGDRIAIISH 1106
Cdd:cd03223 110 PKFVFLDEATSALDEESEDRLYQLL---KELGITVISVGHRPSLWKFHDRVLDLDG 162
|
|
| PRK10247 |
PRK10247 |
putative ABC transporter ATP-binding protein YbbL; Provisional |
1910-2083 |
3.00e-04 |
|
putative ABC transporter ATP-binding protein YbbL; Provisional
Pssm-ID: 182331 [Multi-domain] Cd Length: 225 Bit Score: 44.70 E-value: 3.00e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1910 QNDILEIKELTkiYRRKRKPAVDRICIGIPPGEcFGLL-GVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSILSNIHEVH- 1987
Cdd:PRK10247 4 NSPLLQLQNVG--YLAGDAKILNNISFSLRAGE-FKLItGPSGCGKSTLLKIVASLISPTSGTLLFEGEDISTLKPEIYr 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1988 QNMGYCPQfdaiTELLTG---REHVEFFALLRGVpekevgKVGEWAIRKlGLVKYG------EKYASNYSGGNKRKLSTA 2058
Cdd:PRK10247 81 QQVSYCAQ----TPTLFGdtvYDNLIFPWQIRNQ------QPDPAIFLD-DLERFAlpdtilTKNIAELSGGEKQRISLI 149
|
170 180
....*....|....*....|....*
gi 568925948 2059 MALIGGPPVVFLDEPTTGMDPKARR 2083
Cdd:PRK10247 150 RNLQFMPKVLLLDEITSALDESNKH 174
|
|
| PRK15134 |
PRK15134 |
microcin C ABC transporter ATP-binding protein YejF; Provisional |
1922-2142 |
3.17e-04 |
|
microcin C ABC transporter ATP-binding protein YejF; Provisional
Pssm-ID: 237917 [Multi-domain] Cd Length: 529 Bit Score: 45.85 E-value: 3.17e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1922 IYRRK--RKPAVDRICIGIPPGECFGLLGVNGAGKSTTF-----------KMLTGDTPVTRgdafLNKNSIL---SNIHE 1985
Cdd:PRK15134 291 ILKRTvdHNVVVKNISFTLRPGETLGLVGESGSGKSTTGlallrlinsqgEIWFDGQPLHN----LNRRQLLpvrHRIQV 366
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1986 VHQ--NMGYCPQFDA---ITELLTgrehVEFFALLRGVPEKEVGKVgewaIRKLGL-VKYGEKYASNYSGGNKRKLSTAM 2059
Cdd:PRK15134 367 VFQdpNSSLNPRLNVlqiIEEGLR----VHQPTLSAAQREQQVIAV----MEEVGLdPETRHRYPAEFSGGQRQRIAIAR 438
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2060 ALIGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGR-SVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQHLKNRFG 2138
Cdd:PRK15134 439 ALILKPSLIILDEPTSSLDKTVQAQILALLKSLQQKHQlAYLFISHDLHVVRALCHQVIVLRQGEVVEQGDCERVFAAPQ 518
|
....
gi 568925948 2139 DGYT 2142
Cdd:PRK15134 519 QEYT 522
|
|
| PRK13409 |
PRK13409 |
ribosome biogenesis/translation initiation ATPase RLI; |
1938-2078 |
3.69e-04 |
|
ribosome biogenesis/translation initiation ATPase RLI;
Pssm-ID: 184037 [Multi-domain] Cd Length: 590 Bit Score: 45.57 E-value: 3.69e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGdaflnknSILSNIhevhqNMGYCPQFdaitelLTGREHVEFFALLRG 2017
Cdd:PRK13409 362 IYEGEVIGIVGPNGIGKTTFAKLLAGVLKPDEG-------EVDPEL-----KISYKPQY------IKPDYDGTVEDLLRS 423
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*....
gi 568925948 2018 VPEK--------EVgkvgewaIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMD 2078
Cdd:PRK13409 424 ITDDlgssyyksEI-------IKPLQLERLLDKNVKDLSGGELQRVAIAACLSRDADLYLLDEPSAHLD 485
|
|
| met_CoM_red_A2 |
TIGR03269 |
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in ... |
899-1116 |
3.86e-04 |
|
methyl coenzyme M reductase system, component A2; The enzyme that catalyzes the final step in methanogenesis, methyl coenzyme M reductase, contains alpha, beta, and gamma chains. In older literature, the complex of alpha, beta, and gamma chains was termed component C, while this single chain protein was termed methyl coenzyme M reductase system component A2. [Energy metabolism, Methanogenesis]
Pssm-ID: 132313 [Multi-domain] Cd Length: 520 Bit Score: 45.56 E-value: 3.86e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGL--FPPTSGTA-YILG---KDIRSEMSS-I 971
Cdd:TIGR03269 1 IEVKNLTKKFDG--KEVLKNISFTIEEGEVLGILGRSGAGKSVLMHVLRGMdqYEPTSGRIiYHVAlceKCGYVERPSkV 78
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 972 RQNLGVCPQHNVLFD--------------------ML----------TVEEHIwfyarLKGLSEKHVKAEME-QMALDVg 1020
Cdd:TIGR03269 79 GEPCPVCGGTLEPEEvdfwnlsdklrrrirkriaiMLqrtfalygddTVLDNV-----LEALEEIGYEGKEAvGRAVDL- 152
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1021 LPPSKLKSKTSQ----LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLK--YRQGRTIILSTHHMDEA 1094
Cdd:TIGR03269 153 IEMVQLSHRITHiardLSGGEKQRVVLARQLAKEPFLFLADEPTGTLDPQTAKLVHNALEEavKASGISMVLTSHWPEVI 232
|
250 260
....*....|....*....|..
gi 568925948 1095 DILGDRIAIISHGKLCCVGSSL 1116
Cdd:TIGR03269 233 EDLSDKAIWLENGEIKEEGTPD 254
|
|
| PRK13409 |
PRK13409 |
ribosome biogenesis/translation initiation ATPase RLI; |
889-1104 |
4.77e-04 |
|
ribosome biogenesis/translation initiation ATPase RLI;
Pssm-ID: 184037 [Multi-domain] Cd Length: 590 Bit Score: 45.18 E-value: 4.77e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHlRLGvsiQNLVKVYrdGMKVAVdglalnfyEGQITSFLGHNGAGKTTTMSILTGLFPPtsgtayilgkdirsem 968
Cdd:PRK13409 76 EEEPVH-RYG---VNGFKLY--GLPIPK--------EGKVTGILGPNGIGKTTAVKILSGELIP---------------- 125
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 969 ssirqNLGvCPQHNVlfdmlTVEEHIWFYAR------LKGLSEKHVKAEME-QMaldVGLPPSKLKSKT----------- 1030
Cdd:PRK13409 126 -----NLG-DYEEEP-----SWDEVLKRFRGtelqnyFKKLYNGEIKVVHKpQY---VDLIPKVFKGKVrellkkvderg 191
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1031 -------------------SQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSR----RGIWELLlkyrQGRTIILS 1087
Cdd:PRK13409 192 kldevverlglenildrdiSELSGGELQRVAIAAALLRDADFYFFDEPTSYLDIRQRlnvaRLIRELA----EGKYVLVV 267
|
250
....*....|....*..
gi 568925948 1088 THHMDEADILGDRIAII 1104
Cdd:PRK13409 268 EHDLAVLDYLADNVHIA 284
|
|
| PRK14258 |
PRK14258 |
phosphate ABC transporter ATP-binding protein; Provisional |
2019-2108 |
5.29e-04 |
|
phosphate ABC transporter ATP-binding protein; Provisional
Pssm-ID: 184593 [Multi-domain] Cd Length: 261 Bit Score: 44.26 E-value: 5.29e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2019 PEKEVGKVGEWAIRKLGL---VKYG-EKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDP----KARRFLWNCAL 2090
Cdd:PRK14258 119 PKLEIDDIVESALKDADLwdeIKHKiHKSALDLSGGQQQRLCIARALAVKPKVLLMDEPCFGLDPiasmKVESLIQSLRL 198
|
90
....*....|....*...
gi 568925948 2091 sivKEGRSVVLTSHSMEE 2108
Cdd:PRK14258 199 ---RSELTMVIVSHNLHQ 213
|
|
| PRK11819 |
PRK11819 |
putative ABC transporter ATP-binding protein; Reviewed |
1913-2010 |
7.12e-04 |
|
putative ABC transporter ATP-binding protein; Reviewed
Pssm-ID: 236992 [Multi-domain] Cd Length: 556 Bit Score: 44.72 E-value: 7.12e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1913 ILEIKELTKIYRRKrKPAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTG-DTPVTrGDAFLNKNsilsnihevhQNMG 1991
Cdd:PRK11819 6 IYTMNRVSKVVPPK-KQILKDISLSFFPGAKIGVLGLNGAGKSTLLRIMAGvDKEFE-GEARPAPG----------IKVG 73
|
90
....*....|....*....
gi 568925948 1992 YCPQFDAITELLTGREHVE 2010
Cdd:PRK11819 74 YLPQEPQLDPEKTVRENVE 92
|
|
| SapD |
COG4170 |
ABC-type antimicrobial peptide export system, ATPase component SapD [Defense mechanisms]; |
1930-2136 |
7.48e-04 |
|
ABC-type antimicrobial peptide export system, ATPase component SapD [Defense mechanisms];
Pssm-ID: 443330 [Multi-domain] Cd Length: 331 Bit Score: 44.13 E-value: 7.48e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1930 AVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP----VTRGDAFLNKNSIL------------SNIHEVHQNMGYC 1993
Cdd:COG4170 22 AVDRVSLTLNEGEIRGLVGESGSGKSLIAKAICGITKdnwhVTADRFRWNGIDLLklsprerrkiigREIAMIFQEPSSC 101
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1994 pqFDAITELltGREhveffaLLRGVPEKEVGkvGEW----------AIR---KLGlVKYGEKYASNY----SGGNKRKLS 2056
Cdd:COG4170 102 --LDPSAKI--GDQ------LIEAIPSWTFK--GKWwqrfkwrkkrAIEllhRVG-IKDHKDIMNSYphelTEGECQKVM 168
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2057 TAMALIGGPPVVFLDEPTTGMDPKAR----RFLwnCALSIVKeGRSVVLTSHSMEECEALCTRMAIMVNGRFRCLGSVQH 2132
Cdd:COG4170 169 IAMAIANQPRLLIADEPTNAMESTTQaqifRLL--ARLNQLQ-GTSILLISHDLESISQWADTITVLYCGQTVESGPTEQ 245
|
....
gi 568925948 2133 LKNR 2136
Cdd:COG4170 246 ILKS 249
|
|
| PRK11819 |
PRK11819 |
putative ABC transporter ATP-binding protein; Reviewed |
896-1060 |
8.96e-04 |
|
putative ABC transporter ATP-binding protein; Reviewed
Pssm-ID: 236992 [Multi-domain] Cd Length: 556 Bit Score: 44.34 E-value: 8.96e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 896 RLG---VSIQNLVKVYRDgmKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTaYILGKDIRseMSSIR 972
Cdd:PRK11819 319 RLGdkvIEAENLSKSFGD--RLLIDDLSFSLPPGGIVGIIGPNGAGKSTLFKMITGQEQPDSGT-IKIGETVK--LAYVD 393
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 973 QNlgvcpqHNVLFDMLTVEEHIwfyarLKGLSEKHV-KAEMeqmaldvglpPSKL------------KSKTSQLSGGMQR 1039
Cdd:PRK11819 394 QS------RDALDPNKTVWEEI-----SGGLDIIKVgNREI----------PSRAyvgrfnfkggdqQKKVGVLSGGERN 452
|
170 180
....*....|....*....|.
gi 568925948 1040 KLSVALAFVGGSKVVILDEPT 1060
Cdd:PRK11819 453 RLHLAKTLKQGGNVLLLDEPT 473
|
|
| ABC_UvrA |
cd03238 |
ATP-binding cassette domain of the excision repair protein UvrA; Nucleotide excision repair in ... |
2034-2105 |
1.30e-03 |
|
ATP-binding cassette domain of the excision repair protein UvrA; Nucleotide excision repair in eubacteria is a process that repairs DNA damage by the removal of a 12-13-mer oligonucleotide containing the lesion. Recognition and cleavage of the damaged DNA is a multistep ATP-dependent reaction that requires the UvrA, UvrB, and UvrC proteins. Both UvrA and UvrB are ATPases, with UvrA having two ATP binding sites, which have the characteristic signature of the family of ABC proteins, and UvrB having one ATP binding site that is structurally related to that of helicases.
Pssm-ID: 213205 [Multi-domain] Cd Length: 176 Bit Score: 41.92 E-value: 1.30e-03
10 20 30 40 50 60 70
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....
gi 568925948 2034 LGLVKYGEKyASNYSGGNKRKLSTAMALIGGP-PVVF-LDEPTTGMDPKARRFLWNCALSIVKEGRSVVLTSHS 2105
Cdd:cd03238 76 LGYLTLGQK-LSTLSGGELQRVKLASELFSEPpGTLFiLDEPSTGLHQQDINQLLEVIKGLIDLGNTVILIEHN 148
|
|
| hmuV |
PRK13547 |
heme ABC transporter ATP-binding protein; |
1922-2130 |
1.47e-03 |
|
heme ABC transporter ATP-binding protein;
Pssm-ID: 184132 [Multi-domain] Cd Length: 272 Bit Score: 42.89 E-value: 1.47e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1922 IYRRKRKpAVDRICIGIPPGECFGLLGVNGAGKSTTFKMLTGDTP--------VTRGDAFLNKNSI-------LSNIHEV 1986
Cdd:PRK13547 9 VARRHRA-ILRDLSLRIEPGRVTALLGRNGAGKSTLLKALAGDLTgggaprgaRVTGDVTLNGEPLaaidaprLARLRAV 87
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1987 hqnmgyCPQ-------FDAITELLTGR-EHveffALLRGVPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTA 2058
Cdd:PRK13547 88 ------LPQaaqpafaFSAREIVLLGRyPH----ARRAGALTHRDGEIAWQALALAGATALVGRDVTTLSGGELARVQFA 157
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2059 MAL---------IGGPPVVFLDEPTTGMDPKARRFLWNCALSIVKEGRSVVLT-SHSMEECEALCTRMAIMVNGRFRCLG 2128
Cdd:PRK13547 158 RVLaqlwpphdaAQPPRYLLLDEPTAALDLAHQHRLLDTVRRLARDWNLGVLAiVHDPNLAARHADRIAMLADGAIVAHG 237
|
..
gi 568925948 2129 SV 2130
Cdd:PRK13547 238 AP 239
|
|
| 3a01205 |
TIGR00956 |
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other] |
916-1064 |
1.53e-03 |
|
Pleiotropic Drug Resistance (PDR) Family protein; [Transport and binding proteins, Other]
Pssm-ID: 273362 [Multi-domain] Cd Length: 1394 Bit Score: 43.94 E-value: 1.53e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 916 VDGLAlnfYEGQITSFLGHNGAGKTTTMSILTG-----LFPPTSGTAY--ILGKDIRSEMssiRQNLGVCPQHNVLFDML 988
Cdd:TIGR00956 80 MDGLI---KPGELTVVLGRPGSGCSTLLKTIASntdgfHIGVEGVITYdgITPEEIKKHY---RGDVVYNAETDVHFPHL 153
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 989 TVEEHIWFYARLK-------GLSEKHVKAEMEQMALDV-GLPPSKlKSKTSQ-----LSGGMQRKLSVALAFVGGSKVVI 1055
Cdd:TIGR00956 154 TVGETLDFAARCKtpqnrpdGVSREEYAKHIADVYMATyGLSHTR-NTKVGNdfvrgVSGGERKRVSIAEASLGGAKIQC 232
|
....*....
gi 568925948 1056 LDEPTAGVD 1064
Cdd:TIGR00956 233 WDNATRGLD 241
|
|
| ABC_RNaseL_inhibitor_domain2 |
cd03237 |
The ATP-binding cassette domain 2 of RNase L inhibitor; The ABC ATPase, RNase L inhibitor (RLI) ... |
1941-2082 |
1.55e-03 |
|
The ATP-binding cassette domain 2 of RNase L inhibitor; The ABC ATPase, RNase L inhibitor (RLI), is a key enzyme in ribosomal biogenesis, formation of translation preinitiation complexes, and assembly of HIV capsids. RLI's are not transport proteins and thus cluster with a group of soluble proteins that lack the transmembrane components commonly found in other members of the family. Structurally, RLI's have an N-terminal Fe-S domain and two nucleotide-binding domains which are arranged to form two composite active sites in their interface cleft. RLI is one of the most conserved enzymes between archaea and eukaryotes with a sequence identity of more than 48%. The high degree of evolutionary conservation suggests that RLI performs a central role in archaeal and eukaryotic physiology.
Pssm-ID: 213204 [Multi-domain] Cd Length: 246 Bit Score: 42.78 E-value: 1.55e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1941 GECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNKNSIlsnihevhqnmGYCPQFdaITELLTGRehVEffALLRGVpE 2020
Cdd:cd03237 25 SEVIGILGPNGIGKTTFIKMLAGVLKPDEGDIEIELDTV-----------SYKPQY--IKADYEGT--VR--DLLSSI-T 86
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2021 KEVGKVGEW---AIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKAR 2082
Cdd:cd03237 87 KDFYTHPYFkteIAKPLQIEQILDREVPELSGGELQRVAIAACLSKDADIYLLDEPSAYLDVEQR 151
|
|
| PRK15064 |
PRK15064 |
ABC transporter ATP-binding protein; Provisional |
889-1060 |
2.01e-03 |
|
ABC transporter ATP-binding protein; Provisional
Pssm-ID: 237894 [Multi-domain] Cd Length: 530 Bit Score: 43.34 E-value: 2.01e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 889 EEEPTHlRLGVSIQNLVKVYRDGMkvAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTayilgkdIR-SE 967
Cdd:PRK15064 311 QDKKLH-RNALEVENLTKGFDNGP--LFKNLNLLLEAGERLAIIGENGVGKTTLLRTLVGELEPDSGT-------VKwSE 380
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 968 MSSIrqnlGVCPQ-HNVLFDM-LTVEEHI--WfyaRLKGLSEKHVKAEMEQM---ALDVglppsklKSKTSQLSGGMQRK 1040
Cdd:PRK15064 381 NANI----GYYAQdHAYDFENdLTLFDWMsqW---RQEGDDEQAVRGTLGRLlfsQDDI-------KKSVKVLSGGEKGR 446
|
170 180
....*....|....*....|
gi 568925948 1041 LSVALAFVGGSKVVILDEPT 1060
Cdd:PRK15064 447 MLFGKLMMQKPNVLVMDEPT 466
|
|
| ABCC_SUR2 |
cd03288 |
ATP-binding cassette domain 2 of the sulfonylurea receptor SUR; The SUR domain 2. The ... |
899-1134 |
2.60e-03 |
|
ATP-binding cassette domain 2 of the sulfonylurea receptor SUR; The SUR domain 2. The sulfonylurea receptor SUR is an ATP binding cassette (ABC) protein of the ABCC/MRP family. Unlike other ABC proteins, it has no intrinsic transport function, neither active nor passive, but associates with the potassium channel proteins Kir6.1 or Kir6.2 to form the ATP-sensitive potassium (K(ATP)) channel. Within the channel complex, SUR serves as a regulatory subunit that fine-tunes the gating of Kir6.x in response to alterations in cellular metabolism. It constitutes a major pharmaceutical target as it binds numerous drugs, K(ATP) channel openers and blockers, capable of up- or down-regulating channel activity.
Pssm-ID: 213255 [Multi-domain] Cd Length: 257 Bit Score: 41.82 E-value: 2.60e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 899 VSIQNLVKVYRDGMKVAVDGLALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKDI-RSEMSSIRQNLGV 977
Cdd:cd03288 20 IKIHDLCVRYENNLKPVLKHVKAYIKPGQKVGICGRTGSGKSSLSLAFFRMVDIFDGKIVIDGIDIsKLPLHTLRSRLSI 99
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 978 CPQHNVLFDMLTveehiwfyaRLKGLSEKHVKAEMEQMALDVglppSKLKSKTSQLSGGM--------------QRKL-S 1042
Cdd:cd03288 100 ILQDPILFSGSI---------RFNLDPECKCTDDRLWEALEI----AQLKNMVKSLPGGLdavvteggenfsvgQRQLfC 166
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1043 VALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTH---HMDEADIlgdrIAIISHGKL--CCVGSSLF 1117
Cdd:cd03288 167 LARAFVRKSSILIMDEATASIDMATENILQKVVMTAFADRTVVTIAHrvsTILDADL----VLVLSRGILveCDTPENLL 242
|
250
....*....|....*..
gi 568925948 1118 LKNQlgtGYYLTLVKKD 1134
Cdd:cd03288 243 AQED---GVFASLVRTD 256
|
|
| PRK11000 |
PRK11000 |
maltose/maltodextrin ABC transporter ATP-binding protein MalK; |
1938-2082 |
2.82e-03 |
|
maltose/maltodextrin ABC transporter ATP-binding protein MalK;
Pssm-ID: 182893 [Multi-domain] Cd Length: 369 Bit Score: 42.32 E-value: 2.82e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1938 IPPGECFGLLGVNGAGKSTTFKMLTGDTPVTRGDAFLNkNSILSNIHEVHQNMGYCPQFDAITELLTGREHVEFFALLRG 2017
Cdd:PRK11000 26 IHEGEFVVFVGPSGCGKSTLLRMIAGLEDITSGDLFIG-EKRMNDVPPAERGVGMVFQSYALYPHLSVAENMSFGLKLAG 104
|
90 100 110 120 130 140
....*....|....*....|....*....|....*....|....*....|....*....|....*
gi 568925948 2018 VPEKEVGKVGEWAIRKLGLVKYGEKYASNYSGGNKRKLSTAMALIGGPPVVFLDEPTTGMDPKAR 2082
Cdd:PRK11000 105 AKKEEINQRVNQVAEVLQLAHLLDRKPKALSGGQRQRVAIGRTLVAEPSVFLLDEPLSNLDAALR 169
|
|
| PRK00635 |
PRK00635 |
excinuclease ABC subunit A; Provisional |
1015-1107 |
2.98e-03 |
|
excinuclease ABC subunit A; Provisional
Pssm-ID: 234806 [Multi-domain] Cd Length: 1809 Bit Score: 42.89 E-value: 2.98e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1015 MALDVGLPPSKLKSKTSQLSGGMQRKlsVALAFVGGSKVV----ILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTH 1089
Cdd:PRK00635 459 ILIDLGLPYLTPERALATLSGGEQER--TALAKHLGAELIgityILDEPSIGLHPQDTHKLINVIKKLRdQGNTVLLVEH 536
|
90
....*....|....*....
gi 568925948 1090 hmDEADI-LGDRIAIISHG 1107
Cdd:PRK00635 537 --DEQMIsLADRIIDIGPG 553
|
|
| ABC_Class2 |
cd03227 |
ATP-binding cassette domain of non-transporter proteins; ABC-type Class 2 contains systems ... |
1921-2104 |
3.18e-03 |
|
ATP-binding cassette domain of non-transporter proteins; ABC-type Class 2 contains systems involved in cellular processes other than transport. These families are characterized by the fact that the ABC subunit is made up of duplicated, fused ABC modules (ABC2). No known transmembrane proteins or domains are associated with these proteins.
Pssm-ID: 213194 [Multi-domain] Cd Length: 162 Bit Score: 40.42 E-value: 3.18e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1921 KIYRRKRKPAVDRICIGIPPGECFGLLGVNGAGKSTTFkmltgdtpvtrgdaflnknsilsnihevhqnmgycpqfDAIT 2000
Cdd:cd03227 1 KIVLGRFPSYFVPNDVTFGEGSLTIITGPNGSGKSTIL--------------------------------------DAIG 42
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 2001 ELLTGREHVEFFALLRGVPEKEVGKVGEWAIRKLGLvkygekyasnySGGNKRKLSTAMAL----IGGPPVVFLDEPTTG 2076
Cdd:cd03227 43 LALGGAQSATRRRSGVKAGCIVAAVSAELIFTRLQL-----------SGGEKELSALALILalasLKPRPLYILDEIDRG 111
|
170 180
....*....|....*....|....*...
gi 568925948 2077 MDPKARRFLWNCALSIVKEGRSVVLTSH 2104
Cdd:cd03227 112 LDPRDGQALAEAILEHLVKGAQVIVITH 139
|
|
| PLN03073 |
PLN03073 |
ABC transporter F family; Provisional |
930-1109 |
3.44e-03 |
|
ABC transporter F family; Provisional
Pssm-ID: 215558 [Multi-domain] Cd Length: 718 Bit Score: 42.54 E-value: 3.44e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 930 SFLGHNGAGKTTTMSILTGLFPPTSGTAYilgkdirsemSSIRQNLGVCPQHNVLFDMLTVEEHIWFYARLKGLSEKHVK 1009
Cdd:PLN03073 539 AMVGPNGIGKSTILKLISGELQPSSGTVF----------RSAKVRMAVFSQHHVDGLDLSSNPLLYMMRCFPGVPEQKLR 608
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1010 AEMEQMALDVGLPPSKLKSktsqLSGGmqRKLSVALAFVGGSK--VVILDEPTAGVDPYSRRGIWELLLKYRQGrtIILS 1087
Cdd:PLN03073 609 AHLGSFGVTGNLALQPMYT----LSGG--QKSRVAFAKITFKKphILLLDEPSNHLDLDAVEALIQGLVLFQGG--VLMV 680
|
170 180
....*....|....*....|....
gi 568925948 1088 THhmDEADILG--DRIAIISHGKL 1109
Cdd:PLN03073 681 SH--DEHLISGsvDELWVVSEGKV 702
|
|
| ABC_UvrA |
cd03238 |
ATP-binding cassette domain of the excision repair protein UvrA; Nucleotide excision repair in ... |
1014-1099 |
3.60e-03 |
|
ATP-binding cassette domain of the excision repair protein UvrA; Nucleotide excision repair in eubacteria is a process that repairs DNA damage by the removal of a 12-13-mer oligonucleotide containing the lesion. Recognition and cleavage of the damaged DNA is a multistep ATP-dependent reaction that requires the UvrA, UvrB, and UvrC proteins. Both UvrA and UvrB are ATPases, with UvrA having two ATP binding sites, which have the characteristic signature of the family of ABC proteins, and UvrB having one ATP binding site that is structurally related to that of helicases.
Pssm-ID: 213205 [Multi-domain] Cd Length: 176 Bit Score: 40.77 E-value: 3.60e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1014 QMALDVGLPPSKLKSKTSQLSGG-MQR-KLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHH 1090
Cdd:cd03238 69 QFLIDVGLGYLTLGQKLSTLSGGeLQRvKLASELFSEPPGTLFILDEPSTGLHQQDINQLLEVIKGLIdLGNTVILIEHN 148
|
90
....*....|..
gi 568925948 1091 ---MDEADILGD 1099
Cdd:cd03238 149 ldvLSSADWIID 160
|
|
| CFTR_protein |
TIGR01271 |
cystic fibrosis transmembrane conductor regulator (CFTR); The model describes the cystis ... |
921-1107 |
4.41e-03 |
|
cystic fibrosis transmembrane conductor regulator (CFTR); The model describes the cystis fibrosis transmembrane conductor regulator (CFTR) in eukaryotes. The principal role of this protein is chloride ion conductance. The protein is predicted to consist of 12 transmembrane domains. Mutations or lesions in the genetic loci have been linked to the aetiology of asthma, bronchiectasis, chronic obstructive pulmonary disease etc. Disease-causing mutations have been studied by 36Cl efflux assays in vitro cell cultures and electrophysiology, all of which point to the impairment of chloride channel stability and not the biosynthetic processing per se. [Transport and binding proteins, Anions]
Pssm-ID: 273530 [Multi-domain] Cd Length: 1490 Bit Score: 42.59 E-value: 4.41e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 921 LNFY--EGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKdirsemssirqnLGVCPQHNVLFDMlTVEEHIWFya 998
Cdd:TIGR01271 445 ISFKleKGQLLAVAGSTGSGKSSLLMMIMGELEPSEGKIKHSGR------------ISFSPQTSWIMPG-TIKDNIIF-- 509
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 999 rlkGLSEKH------VKAEmeQMALDVGLPPSKLKSKTSQ----LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSR 1068
Cdd:TIGR01271 510 ---GLSYDEyrytsvIKAC--QLEEDIALFPEKDKTVLGEggitLSGGQRARISLARAVYKDADLYLLDSPFTHLDVVTE 584
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 568925948 1069 RGIWE-LLLKYRQGRTIILST---HHMDEAdilgDRIAIISHG 1107
Cdd:TIGR01271 585 KEIFEsCLCKLMSNKTRILVTsklEHLKKA----DKILLLHEG 623
|
|
| uvra |
TIGR00630 |
excinuclease ABC, A subunit; This family is a member of the ABC transporter superfamily of ... |
981-1092 |
5.93e-03 |
|
excinuclease ABC, A subunit; This family is a member of the ABC transporter superfamily of proteins of which all members for which functions are known except the UvrA proteins are involved in the transport of material through membranes. UvrA orthologs are involved in the recognition of DNA damage as a step in nucleotide excision repair. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair]
Pssm-ID: 273184 [Multi-domain] Cd Length: 925 Bit Score: 41.92 E-value: 5.93e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 981 HNVLfDMlTVEEHIWFYARLKGLSEKHvkaemeQMALDVGLPPSKLKSKTSQLSGG-MQR-KLSVAL-AFVGGSKVVILD 1057
Cdd:TIGR00630 786 ADVL-DM-TVEEAYEFFEAVPSISRKL------QTLCDVGLGYIRLGQPATTLSGGeAQRiKLAKELsKRSTGRTLYILD 857
|
90 100 110
....*....|....*....|....*....|....*.
gi 568925948 1058 EPTAGVDPYSRRGIWELLLKYR-QGRTIILSTHHMD 1092
Cdd:TIGR00630 858 EPTTGLHFDDIKKLLEVLQRLVdKGNTVVVIEHNLD 893
|
|
| PRK15093 |
PRK15093 |
peptide ABC transporter ATP-binding protein SapD; |
1019-1108 |
8.32e-03 |
|
peptide ABC transporter ATP-binding protein SapD;
Pssm-ID: 185049 [Multi-domain] Cd Length: 330 Bit Score: 40.94 E-value: 8.32e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1019 VGLPPSK--LKSKTSQLSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRGIWELLLKYRQ--GRTIILSTHHMDEA 1094
Cdd:PRK15093 143 VGIKDHKdaMRSFPYELTEGECQKVMIAIALANQPRLLIADEPTNAMEPTTQAQIFRLLTRLNQnnNTTILLISHDLQML 222
|
90
....*....|....
gi 568925948 1095 DILGDRIAIISHGK 1108
Cdd:PRK15093 223 SQWADKINVLYCGQ 236
|
|
| ABCC_CFTR1 |
cd03291 |
ATP-binding cassette domain of the cystic fibrosis transmembrane regulator, subfamily C; The ... |
919-1107 |
8.38e-03 |
|
ATP-binding cassette domain of the cystic fibrosis transmembrane regulator, subfamily C; The CFTR subfamily domain 1. The cystic fibrosis transmembrane regulator (CFTR), the product of the gene mutated in patients with cystic fibrosis, has adapted the ABC transporter structural motif to form a tightly regulated anion channel at the apical surface of many epithelia. Use of the term assembly of a functional ion channel implies the coming together of subunits, or at least smaller not-yet functional components of the active whole. In fact, on the basis of current knowledge only the CFTR polypeptide itself is required to form an ATP- and protein kinase A-dependent low-conductance chloride channel of the type present in the apical membrane of many epithelial cells. CFTR displays the typical organization (IM-ABC)2 and carries a characteristic hydrophilic R-domain that separates IM1-ABC1 from IM2-ABC2.
Pssm-ID: 213258 [Multi-domain] Cd Length: 282 Bit Score: 40.61 E-value: 8.38e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 919 LALNFYEGQITSFLGHNGAGKTTTMSILTGLFPPTSGTAYILGKdirsemssirqnLGVCPQHNVLFDMlTVEEHIWFya 998
Cdd:cd03291 56 INLKIEKGEMLAITGSTGSGKTSLLMLILGELEPSEGKIKHSGR------------ISFSSQFSWIMPG-TIKENIIF-- 120
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 999 rlkGLS--EKHVKAEME--QMALDVGLPPSKLKSKTSQ----LSGGMQRKLSVALAFVGGSKVVILDEPTAGVDPYSRRG 1070
Cdd:cd03291 121 ---GVSydEYRYKSVVKacQLEEDITKFPEKDNTVLGEggitLSGGQRARISLARAVYKDADLYLLDSPFGYLDVFTEKE 197
|
170 180 190
....*....|....*....|....*....|....*...
gi 568925948 1071 IWE-LLLKYRQGRTIILSTHHMDEADIlGDRIAIISHG 1107
Cdd:cd03291 198 IFEsCVCKLMANKTRILVTSKMEHLKK-ADKILILHEG 234
|
|
| YhaN |
COG4717 |
Uncharacterized conserved protein YhaN, contains AAA domain [Function unknown]; |
1032-1106 |
8.70e-03 |
|
Uncharacterized conserved protein YhaN, contains AAA domain [Function unknown];
Pssm-ID: 443752 [Multi-domain] Cd Length: 641 Bit Score: 41.29 E-value: 8.70e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 568925948 1032 QLSGG--MQRKLSVALAFV---GGSKV-VILDEPTAGVDPYSRRGIWELLLKYRQGRTIILSTHHMDEADILGDRIAIIS 1105
Cdd:COG4717 558 ELSRGtrEQLYLALRLALAellAGEPLpLILDDAFVNFDDERLRAALELLAELAKGRQVIYFTCHEELVELFQEEGAHVI 637
|
.
gi 568925948 1106 H 1106
Cdd:COG4717 638 E 638
|
|
| |