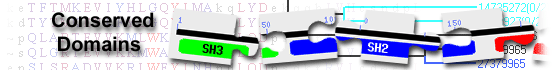|
Name |
Accession |
Description |
Interval |
E-value |
| Nup153 |
pfam08604 |
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and ... |
113-621 |
0e+00 |
|
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and Nup154 from fission yeast. These have been demonstrated to be functionally equivalent.
:
Pssm-ID: 462533 [Multi-domain] Cd Length: 507 Bit Score: 740.16 E-value: 0e+00
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 113 PSTTSTASNYPDVLTRPSLHRSHLNFSMLESPALHCQPSTSSAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 192
Cdd:pfam08604 1 PSTSRSALNFQDVLTRPSLHRSHLNFTVLDSPALFCQPSTSTAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 193 DKDITVSKNTSLPPLWSPEAERSHSLSQHTATSSKKPAFNLSAFGTLSPSLGNSSILKTSQLGDSPFYPGKTTYGGaAAA 272
Cdd:pfam08604 81 DKDIPVSKTVSLPLLWSPEAERSHSLSQHSASSSKKPAFNLSAFGTLSPSLGNSSVLNTSQLGDSPFYPGKTTYGG-AAA 159
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 273 VRQSKLRNTPYQAPVRRQMKAKQLSAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSIVSSPLNSPLDRSGIDITDFQA 352
Cdd:pfam08604 160 VRSSRVRGTPYQAPVRRQIKAKPANAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSTVSSPLSSPLDRSDLDITNFQS 239
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 353 KREKVDSQYPPVQRLMTPKPVSIATNRSVYFKPSLTPSGEFRKTNQRIDNKCSTgYEKNMTPGQNREQRESGFSYPNFSL 432
Cdd:pfam08604 240 KRKKVDSQYPPVQKLVTPKAISVSGNRSVYFKPSLTPSGQSNKTSRRVDKDHRE-TEVRQSPEQAVEPSESSLSYPKFST 318
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 433 PAANGLSSGVGGGGGKMRRERTRFVASKPLEEEE-MEVPVLPKISLPITSSSLPTFNFSSPEITTSSPSPINSSQALTNK 511
Cdd:pfam08604 319 PASNSLSSGGNGGGGKMRRERGRRPSSKPAEEDEvVEVPVLPKISLPISTASLPTFNFSSPTSSTSSSSPPSVTKSAENK 398
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 512 VQMTSPSSTGSPMFKFSSPIVKSTEANVLPP-SSIGFTFSVPVAKTAELSGSSSTLEPIISSSAHhvTTVNSTNCKKTPP 590
Cdd:pfam08604 399 AQKQPPSTSGSPAFTFSSPIVKSTEASVPPPsPSIGFTFSVPVAKTAELSGSSGTSVVSVVSTET--TTVNSTSNKKEEE 476
|
490 500 510
....*....|....*....|....*....|.
gi 24430146 591 EDCEGPFRPAEILKEGSVLDILKSPGFASPK 621
Cdd:pfam08604 477 EEFEGPFKPAKVLKQGSVLDILKSPGFASPK 507
|
|
| Nup_retrotrp_bd |
pfam10599 |
Retro-transposon transporting motif; This is the highly conserved C-terminal motif ... |
1386-1475 |
2.90e-18 |
|
Retro-transposon transporting motif; This is the highly conserved C-terminal motif GRKIxxxxxRRKx of nucleoporins that plays a critical and unique role in the nuclear import of retro-transposons in both yeasts and higher organizms. It would appear that the arginine residues at positions 2 and 9-10 constitute a bipartite nuclear localization signal, with two basic peptide motifs separated by an interchangeable spacer sequence, that is crucial for the retro-transposon activity.
:
Pssm-ID: 371156 Cd Length: 86 Bit Score: 80.88 E-value: 2.90e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1386 GTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGANSSTPAASAQPSGSGGFPFNQSPAAFTVGSNGKNvfsssGTSFSGR 1465
Cdd:pfam10599 2 EGTPDPASVFGFGSSNTNFNFTNNNPSTPFGFGASPSTPQAANPPSAFTPPAFSQSPASFNVGSNGSN-----MFSVSGR 76
|
90
....*....|
gi 24430146 1466 KIKTAVRRRK 1475
Cdd:pfam10599 77 KIAQMRRRRR 86
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
723-751 |
2.17e-09 |
|
Zn-finger in Ran binding protein and others;
:
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 53.90 E-value: 2.17e-09
10 20
....*....|....*....|....*....
gi 24430146 723 IGTWDCDTCLVQNKPEAIKCVACETPKPG 751
Cdd:pfam00641 2 EGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
851-880 |
8.69e-09 |
|
Zn-finger in Ran binding protein and others;
:
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 52.35 E-value: 8.69e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 851 PEGSWDCELCLVQNKADSTKCLACESAKPG 880
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
793-822 |
9.87e-09 |
|
Zn-finger in Ran binding protein and others;
:
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 51.97 E-value: 9.87e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 793 PIGSWECSVCCVSNNAEDNKCVSCMSEKPG 822
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
661-687 |
3.58e-07 |
|
Zn-finger in Ran binding protein and others;
:
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 47.73 E-value: 3.58e-07
10 20
....*....|....*....|....*..
gi 24430146 661 SWQCDTCLLQNKVTDNKCIACQAAKLS 687
Cdd:pfam00641 4 DWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| Chi1 super family |
cl43877 |
Chitinase [Carbohydrate transport and metabolism]; |
1261-1437 |
3.78e-05 |
|
Chitinase [Carbohydrate transport and metabolism];
The actual alignment was detected with superfamily member COG3469:
Pssm-ID: 442692 [Multi-domain] Cd Length: 534 Bit Score: 48.21 E-value: 3.78e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1261 ATTSSTGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSSFVFGTGPSAPSASPAFGANQTPTFGQSQGASQPNPPG 1340
Cdd:COG3469 37 ATATTVVSTTGSVVVAASGSAGSGTGTTAASSTAATSSTTSTTATATAAAAAATSTSATLVATSTASGANTGTSTVTTTS 116
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1341 FGSISSSTALFPTGSQPAPPTFGTVSSSSQPPVFGQQPSQSAFGSGTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGAN 1420
Cdd:COG3469 117 TGAGSVTSTTSSTAGSTTTSGASATSSAGSTTTTTTVSGTETATGGTTTTSTTTTTTSASTTPSATTTATATTASGATTP 196
|
170
....*....|....*..
gi 24430146 1421 SSTPAASAQPSGSGGFP 1437
Cdd:COG3469 197 SATTTATTTGPPTPGLP 213
|
|
| |
|
Name |
Accession |
Description |
Interval |
E-value |
| Nup153 |
pfam08604 |
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and ... |
113-621 |
0e+00 |
|
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and Nup154 from fission yeast. These have been demonstrated to be functionally equivalent.
Pssm-ID: 462533 [Multi-domain] Cd Length: 507 Bit Score: 740.16 E-value: 0e+00
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 113 PSTTSTASNYPDVLTRPSLHRSHLNFSMLESPALHCQPSTSSAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 192
Cdd:pfam08604 1 PSTSRSALNFQDVLTRPSLHRSHLNFTVLDSPALFCQPSTSTAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 193 DKDITVSKNTSLPPLWSPEAERSHSLSQHTATSSKKPAFNLSAFGTLSPSLGNSSILKTSQLGDSPFYPGKTTYGGaAAA 272
Cdd:pfam08604 81 DKDIPVSKTVSLPLLWSPEAERSHSLSQHSASSSKKPAFNLSAFGTLSPSLGNSSVLNTSQLGDSPFYPGKTTYGG-AAA 159
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 273 VRQSKLRNTPYQAPVRRQMKAKQLSAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSIVSSPLNSPLDRSGIDITDFQA 352
Cdd:pfam08604 160 VRSSRVRGTPYQAPVRRQIKAKPANAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSTVSSPLSSPLDRSDLDITNFQS 239
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 353 KREKVDSQYPPVQRLMTPKPVSIATNRSVYFKPSLTPSGEFRKTNQRIDNKCSTgYEKNMTPGQNREQRESGFSYPNFSL 432
Cdd:pfam08604 240 KRKKVDSQYPPVQKLVTPKAISVSGNRSVYFKPSLTPSGQSNKTSRRVDKDHRE-TEVRQSPEQAVEPSESSLSYPKFST 318
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 433 PAANGLSSGVGGGGGKMRRERTRFVASKPLEEEE-MEVPVLPKISLPITSSSLPTFNFSSPEITTSSPSPINSSQALTNK 511
Cdd:pfam08604 319 PASNSLSSGGNGGGGKMRRERGRRPSSKPAEEDEvVEVPVLPKISLPISTASLPTFNFSSPTSSTSSSSPPSVTKSAENK 398
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 512 VQMTSPSSTGSPMFKFSSPIVKSTEANVLPP-SSIGFTFSVPVAKTAELSGSSSTLEPIISSSAHhvTTVNSTNCKKTPP 590
Cdd:pfam08604 399 AQKQPPSTSGSPAFTFSSPIVKSTEASVPPPsPSIGFTFSVPVAKTAELSGSSGTSVVSVVSTET--TTVNSTSNKKEEE 476
|
490 500 510
....*....|....*....|....*....|.
gi 24430146 591 EDCEGPFRPAEILKEGSVLDILKSPGFASPK 621
Cdd:pfam08604 477 EEFEGPFKPAKVLKQGSVLDILKSPGFASPK 507
|
|
| Nup_retrotrp_bd |
pfam10599 |
Retro-transposon transporting motif; This is the highly conserved C-terminal motif ... |
1386-1475 |
2.90e-18 |
|
Retro-transposon transporting motif; This is the highly conserved C-terminal motif GRKIxxxxxRRKx of nucleoporins that plays a critical and unique role in the nuclear import of retro-transposons in both yeasts and higher organizms. It would appear that the arginine residues at positions 2 and 9-10 constitute a bipartite nuclear localization signal, with two basic peptide motifs separated by an interchangeable spacer sequence, that is crucial for the retro-transposon activity.
Pssm-ID: 371156 Cd Length: 86 Bit Score: 80.88 E-value: 2.90e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1386 GTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGANSSTPAASAQPSGSGGFPFNQSPAAFTVGSNGKNvfsssGTSFSGR 1465
Cdd:pfam10599 2 EGTPDPASVFGFGSSNTNFNFTNNNPSTPFGFGASPSTPQAANPPSAFTPPAFSQSPASFNVGSNGSN-----MFSVSGR 76
|
90
....*....|
gi 24430146 1466 KIKTAVRRRK 1475
Cdd:pfam10599 77 KIAQMRRRRR 86
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
723-751 |
2.17e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 53.90 E-value: 2.17e-09
10 20
....*....|....*....|....*....
gi 24430146 723 IGTWDCDTCLVQNKPEAIKCVACETPKPG 751
Cdd:pfam00641 2 EGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
851-880 |
8.69e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 52.35 E-value: 8.69e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 851 PEGSWDCELCLVQNKADSTKCLACESAKPG 880
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
793-822 |
9.87e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 51.97 E-value: 9.87e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 793 PIGSWECSVCCVSNNAEDNKCVSCMSEKPG 822
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
661-687 |
3.58e-07 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 47.73 E-value: 3.58e-07
10 20
....*....|....*....|....*..
gi 24430146 661 SWQCDTCLLQNKVTDNKCIACQAAKLS 687
Cdd:pfam00641 4 DWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
724-748 |
7.86e-06 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 43.85 E-value: 7.86e-06
|
| Chi1 |
COG3469 |
Chitinase [Carbohydrate transport and metabolism]; |
1261-1437 |
3.78e-05 |
|
Chitinase [Carbohydrate transport and metabolism];
Pssm-ID: 442692 [Multi-domain] Cd Length: 534 Bit Score: 48.21 E-value: 3.78e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1261 ATTSSTGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSSFVFGTGPSAPSASPAFGANQTPTFGQSQGASQPNPPG 1340
Cdd:COG3469 37 ATATTVVSTTGSVVVAASGSAGSGTGTTAASSTAATSSTTSTTATATAAAAAATSTSATLVATSTASGANTGTSTVTTTS 116
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1341 FGSISSSTALFPTGSQPAPPTFGTVSSSSQPPVFGQQPSQSAFGSGTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGAN 1420
Cdd:COG3469 117 TGAGSVTSTTSSTAGSTTTSGASATSSAGSTTTTTTVSGTETATGGTTTTSTTTTTTSASTTPSATTTATATTASGATTP 196
|
170
....*....|....*..
gi 24430146 1421 SSTPAASAQPSGSGGFP 1437
Cdd:COG3469 197 SATTTATTTGPPTPGLP 213
|
|
| Nucleoporin_FG2 |
pfam15967 |
Nucleoporin FG repeated region; Nucleoporin_FG2, or nucleoporin p58/p45, is a family of ... |
1265-1441 |
2.14e-04 |
|
Nucleoporin FG repeated region; Nucleoporin_FG2, or nucleoporin p58/p45, is a family of chordate nucleoporins. The proteins carry many repeats of the FG sequence motif.
Pssm-ID: 435043 [Multi-domain] Cd Length: 586 Bit Score: 45.81 E-value: 2.14e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1265 STGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSS--------------------FVFGTgPSAPSASPAFGANQT 1324
Cdd:pfam15967 12 STATAGGGFSFGAAAASNPGSTGGFSFGTLGAAPAATATtttatlglggglfgqkpatgFTFGT-PASSTAATGPTGLTL 90
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1325 PTFGQSQGASQPNPPGFGSISSST---ALFPTGSQPAPPTFGTVSSSSQPPvfgQQPSQSAFGSGTTPNSSSAFQFG--- 1398
Cdd:pfam15967 91 GTPAATTAASTGFSLGFNKPAASAtpfSLPASSTSGGGLSLGSVLTSTAAQ---QGATGFTLNLGGTPATTTAVSTGlsl 167
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 24430146 1399 ----SSTTNFNFTNNSPSGV------FTFGANSSTPAASAQPS-GSGGFPFNQS 1441
Cdd:pfam15967 168 gstlTSLGGSLFQNTNSTGLgqttlgLTLLATSTAPVSAPAASeGLGGLDFSTS 221
|
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
660-683 |
2.63e-04 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 39.61 E-value: 2.63e-04
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
853-875 |
3.26e-04 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 39.22 E-value: 3.26e-04
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
795-816 |
1.49e-03 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 37.30 E-value: 1.49e-03
|
| |
|
Name |
Accession |
Description |
Interval |
E-value |
| Nup153 |
pfam08604 |
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and ... |
113-621 |
0e+00 |
|
Nucleoporin Nup153-like; This family contains both the nucleoporin Nup153 from human and Nup154 from fission yeast. These have been demonstrated to be functionally equivalent.
Pssm-ID: 462533 [Multi-domain] Cd Length: 507 Bit Score: 740.16 E-value: 0e+00
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 113 PSTTSTASNYPDVLTRPSLHRSHLNFSMLESPALHCQPSTSSAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 192
Cdd:pfam08604 1 PSTSRSALNFQDVLTRPSLHRSHLNFTVLDSPALFCQPSTSTAFPIGSSGFSLVKEIKDSTSQHDDDNISTTSGFSSRAS 80
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 193 DKDITVSKNTSLPPLWSPEAERSHSLSQHTATSSKKPAFNLSAFGTLSPSLGNSSILKTSQLGDSPFYPGKTTYGGaAAA 272
Cdd:pfam08604 81 DKDIPVSKTVSLPLLWSPEAERSHSLSQHSASSSKKPAFNLSAFGTLSPSLGNSSVLNTSQLGDSPFYPGKTTYGG-AAA 159
|
170 180 190 200 210 220 230 240
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 273 VRQSKLRNTPYQAPVRRQMKAKQLSAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSIVSSPLNSPLDRSGIDITDFQA 352
Cdd:pfam08604 160 VRSSRVRGTPYQAPVRRQIKAKPANAQSYGVTSSTARRILQSLEKMSSPLADAKRIPSTVSSPLSSPLDRSDLDITNFQS 239
|
250 260 270 280 290 300 310 320
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 353 KREKVDSQYPPVQRLMTPKPVSIATNRSVYFKPSLTPSGEFRKTNQRIDNKCSTgYEKNMTPGQNREQRESGFSYPNFSL 432
Cdd:pfam08604 240 KRKKVDSQYPPVQKLVTPKAISVSGNRSVYFKPSLTPSGQSNKTSRRVDKDHRE-TEVRQSPEQAVEPSESSLSYPKFST 318
|
330 340 350 360 370 380 390 400
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 433 PAANGLSSGVGGGGGKMRRERTRFVASKPLEEEE-MEVPVLPKISLPITSSSLPTFNFSSPEITTSSPSPINSSQALTNK 511
Cdd:pfam08604 319 PASNSLSSGGNGGGGKMRRERGRRPSSKPAEEDEvVEVPVLPKISLPISTASLPTFNFSSPTSSTSSSSPPSVTKSAENK 398
|
410 420 430 440 450 460 470 480
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 512 VQMTSPSSTGSPMFKFSSPIVKSTEANVLPP-SSIGFTFSVPVAKTAELSGSSSTLEPIISSSAHhvTTVNSTNCKKTPP 590
Cdd:pfam08604 399 AQKQPPSTSGSPAFTFSSPIVKSTEASVPPPsPSIGFTFSVPVAKTAELSGSSGTSVVSVVSTET--TTVNSTSNKKEEE 476
|
490 500 510
....*....|....*....|....*....|.
gi 24430146 591 EDCEGPFRPAEILKEGSVLDILKSPGFASPK 621
Cdd:pfam08604 477 EEFEGPFKPAKVLKQGSVLDILKSPGFASPK 507
|
|
| Nup_retrotrp_bd |
pfam10599 |
Retro-transposon transporting motif; This is the highly conserved C-terminal motif ... |
1386-1475 |
2.90e-18 |
|
Retro-transposon transporting motif; This is the highly conserved C-terminal motif GRKIxxxxxRRKx of nucleoporins that plays a critical and unique role in the nuclear import of retro-transposons in both yeasts and higher organizms. It would appear that the arginine residues at positions 2 and 9-10 constitute a bipartite nuclear localization signal, with two basic peptide motifs separated by an interchangeable spacer sequence, that is crucial for the retro-transposon activity.
Pssm-ID: 371156 Cd Length: 86 Bit Score: 80.88 E-value: 2.90e-18
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1386 GTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGANSSTPAASAQPSGSGGFPFNQSPAAFTVGSNGKNvfsssGTSFSGR 1465
Cdd:pfam10599 2 EGTPDPASVFGFGSSNTNFNFTNNNPSTPFGFGASPSTPQAANPPSAFTPPAFSQSPASFNVGSNGSN-----MFSVSGR 76
|
90
....*....|
gi 24430146 1466 KIKTAVRRRK 1475
Cdd:pfam10599 77 KIAQMRRRRR 86
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
723-751 |
2.17e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 53.90 E-value: 2.17e-09
10 20
....*....|....*....|....*....
gi 24430146 723 IGTWDCDTCLVQNKPEAIKCVACETPKPG 751
Cdd:pfam00641 2 EGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
851-880 |
8.69e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 52.35 E-value: 8.69e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 851 PEGSWDCELCLVQNKADSTKCLACESAKPG 880
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
793-822 |
9.87e-09 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 51.97 E-value: 9.87e-09
10 20 30
....*....|....*....|....*....|
gi 24430146 793 PIGSWECSVCCVSNNAEDNKCVSCMSEKPG 822
Cdd:pfam00641 1 REGDWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| zf-RanBP |
pfam00641 |
Zn-finger in Ran binding protein and others; |
661-687 |
3.58e-07 |
|
Zn-finger in Ran binding protein and others;
Pssm-ID: 395516 [Multi-domain] Cd Length: 30 Bit Score: 47.73 E-value: 3.58e-07
10 20
....*....|....*....|....*..
gi 24430146 661 SWQCDTCLLQNKVTDNKCIACQAAKLS 687
Cdd:pfam00641 4 DWDCSKCLVQNFATSTKCVACQAPKPD 30
|
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
724-748 |
7.86e-06 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 43.85 E-value: 7.86e-06
|
| Chi1 |
COG3469 |
Chitinase [Carbohydrate transport and metabolism]; |
1261-1437 |
3.78e-05 |
|
Chitinase [Carbohydrate transport and metabolism];
Pssm-ID: 442692 [Multi-domain] Cd Length: 534 Bit Score: 48.21 E-value: 3.78e-05
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1261 ATTSSTGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSSFVFGTGPSAPSASPAFGANQTPTFGQSQGASQPNPPG 1340
Cdd:COG3469 37 ATATTVVSTTGSVVVAASGSAGSGTGTTAASSTAATSSTTSTTATATAAAAAATSTSATLVATSTASGANTGTSTVTTTS 116
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1341 FGSISSSTALFPTGSQPAPPTFGTVSSSSQPPVFGQQPSQSAFGSGTTPNSSSAFQFGSSTTNFNFTNNSPSGVFTFGAN 1420
Cdd:COG3469 117 TGAGSVTSTTSSTAGSTTTSGASATSSAGSTTTTTTVSGTETATGGTTTTSTTTTTTSASTTPSATTTATATTASGATTP 196
|
170
....*....|....*..
gi 24430146 1421 SSTPAASAQPSGSGGFP 1437
Cdd:COG3469 197 SATTTATTTGPPTPGLP 213
|
|
| Nucleoporin_FG2 |
pfam15967 |
Nucleoporin FG repeated region; Nucleoporin_FG2, or nucleoporin p58/p45, is a family of ... |
1265-1441 |
2.14e-04 |
|
Nucleoporin FG repeated region; Nucleoporin_FG2, or nucleoporin p58/p45, is a family of chordate nucleoporins. The proteins carry many repeats of the FG sequence motif.
Pssm-ID: 435043 [Multi-domain] Cd Length: 586 Bit Score: 45.81 E-value: 2.14e-04
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1265 STGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSS--------------------FVFGTgPSAPSASPAFGANQT 1324
Cdd:pfam15967 12 STATAGGGFSFGAAAASNPGSTGGFSFGTLGAAPAATATtttatlglggglfgqkpatgFTFGT-PASSTAATGPTGLTL 90
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1325 PTFGQSQGASQPNPPGFGSISSST---ALFPTGSQPAPPTFGTVSSSSQPPvfgQQPSQSAFGSGTTPNSSSAFQFG--- 1398
Cdd:pfam15967 91 GTPAATTAASTGFSLGFNKPAASAtpfSLPASSTSGGGLSLGSVLTSTAAQ---QGATGFTLNLGGTPATTTAVSTGlsl 167
|
170 180 190 200 210
....*....|....*....|....*....|....*....|....*....|....
gi 24430146 1399 ----SSTTNFNFTNNSPSGV------FTFGANSSTPAASAQPS-GSGGFPFNQS 1441
Cdd:pfam15967 168 gstlTSLGGSLFQNTNSTGLgqttlgLTLLATSTAPVSAPAASeGLGGLDFSTS 221
|
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
660-683 |
2.63e-04 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 39.61 E-value: 2.63e-04
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
853-875 |
3.26e-04 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 39.22 E-value: 3.26e-04
|
| ZnF_RBZ |
smart00547 |
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. ... |
795-816 |
1.49e-03 |
|
Zinc finger domain; Zinc finger domain in Ran-binding proteins (RanBPs), and other proteins. In RanBPs, this domain binds RanGDP.
Pssm-ID: 197784 [Multi-domain] Cd Length: 25 Bit Score: 37.30 E-value: 1.49e-03
|
| Chi1 |
COG3469 |
Chitinase [Carbohydrate transport and metabolism]; |
1208-1394 |
3.83e-03 |
|
Chitinase [Carbohydrate transport and metabolism];
Pssm-ID: 442692 [Multi-domain] Cd Length: 534 Bit Score: 41.66 E-value: 3.83e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1208 GGGIFGSSTSSSNPPVATFVFGQSSNPVSSSAFGNTAESSTSQSLL----FSQDSKLATTSSTGTAVTPFVFGPGASSNN 1283
Cdd:COG3469 24 GAAATAASVTLTAATATTVVSTTGSVVVAASGSAGSGTGTTAASSTaatsSTTSTTATATAAAAAATSTSATLVATSTAS 103
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1284 TTTSGFGFGATTTSSSAGssfvfGTGPSAPSASPAFGANQTPTFGQSQGASQPNPPG--FGSISSSTALFPTGSQPAPPT 1361
Cdd:COG3469 104 GANTGTSTVTTTSTGAGS-----VTSTTSSTAGSTTTSGASATSSAGSTTTTTTVSGteTATGGTTTTSTTTTTTSASTT 178
|
170 180 190
....*....|....*....|....*....|...
gi 24430146 1362 FGTVSSSSQPPVFGQQPSQSAFGSGTTPNSSSA 1394
Cdd:COG3469 179 PSATTTATATTASGATTPSATTTATTTGPPTPG 211
|
|
| Chi1 |
COG3469 |
Chitinase [Carbohydrate transport and metabolism]; |
1174-1372 |
7.52e-03 |
|
Chitinase [Carbohydrate transport and metabolism];
Pssm-ID: 442692 [Multi-domain] Cd Length: 534 Bit Score: 40.51 E-value: 7.52e-03
10 20 30 40 50 60 70 80
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1174 GAQTSTTADQGAAKPVFSFLNNSSSSSSTPATSAGG-GIFGSSTSSSNPPVATFVFGQSSNPVSSSAFG-NTAESSTSQS 1251
Cdd:COG3469 12 AGGASATAVTLLGAAATAASVTLTAATATTVVSTTGsVVVAASGSAGSGTGTTAASSTAATSSTTSTTAtATAAAAAATS 91
|
90 100 110 120 130 140 150 160
....*....|....*....|....*....|....*....|....*....|....*....|....*....|....*....|
gi 24430146 1252 LLFSQDSKLATTSSTGTAVTPFVFGPGASSNNTTTSGFGFGATTTSSSAGSSFVFGTGPSAPSASPAF--GANQTPTFGq 1329
Cdd:COG3469 92 TSATLVATSTASGANTGTSTVTTTSTGAGSVTSTTSSTAGSTTTSGASATSSAGSTTTTTTVSGTETAtgGTTTTSTTT- 170
|
170 180 190 200
....*....|....*....|....*....|....*....|...
gi 24430146 1330 sQGASQPNPPGFGSISSSTALFPTGSQPAPPTFGTVSSSSQPP 1372
Cdd:COG3469 171 -TTTSASTTPSATTTATATTASGATTPSATTTATTTGPPTPGL 212
|
|
| |