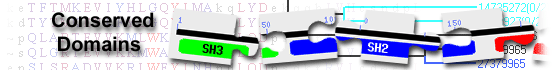integrin alpha-5 precursor [Homo sapiens]
integrin alpha( domain architecture ID 12192630)
integrin alpha is a transmembrane receptor that facilitates cell-extracellular matrix (ECM) adhesion
List of domain hits

| Name | Accession | Description | Interval | E-value | |||||||
| Integrin_alpha2 | pfam08441 | Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C ... |
490-921 | 0e+00 | |||||||
Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C terminus of a number of pfam01839 repeats and to the N-terminus of the pfam00357 cytoplasmic region. This region is composed of three immunoglobulin-like domains. : Pssm-ID: 462478 [Multi-domain] Cd Length: 449 Bit Score: 560.40 E-value: 0e+00
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
389-443 | 3.43e-14 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. : Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 67.79 E-value: 3.43e-14
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
322-379 | 1.34e-12 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. : Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 63.16 E-value: 1.34e-12
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
56-114 | 3.74e-08 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. : Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 50.84 E-value: 3.74e-08
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
455-498 | 1.14e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. : Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 43.90 E-value: 1.14e-05
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
268-310 | 8.53e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. : Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 41.21 E-value: 8.53e-05
|
|||||||||||
| Name | Accession | Description | Interval | E-value | |||||||
| Integrin_alpha2 | pfam08441 | Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C ... |
490-921 | 0e+00 | |||||||
Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C terminus of a number of pfam01839 repeats and to the N-terminus of the pfam00357 cytoplasmic region. This region is composed of three immunoglobulin-like domains. Pssm-ID: 462478 [Multi-domain] Cd Length: 449 Bit Score: 560.40 E-value: 0e+00
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
389-443 | 3.43e-14 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 67.79 E-value: 3.43e-14
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
322-379 | 1.34e-12 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 63.16 E-value: 1.34e-12
|
|||||||||||
| FG-GAP | pfam01839 | FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven ... |
326-368 | 1.00e-09 | |||||||
FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven copies in alpha integrins. This repeat has been predicted to fold into a beta propeller structure. The repeat is called the FG-GAP repeat after two conserved motifs in the repeat. The FG-GAP repeats are found in the N terminus of integrin alpha chains, a region that has been shown to be important for ligand binding. A putative Ca2+ binding motif is found in some of the repeats. Pssm-ID: 460357 Cd Length: 36 Bit Score: 54.44 E-value: 1.00e-09
|
|||||||||||
| FG-GAP | pfam01839 | FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven ... |
392-428 | 1.58e-08 | |||||||
FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven copies in alpha integrins. This repeat has been predicted to fold into a beta propeller structure. The repeat is called the FG-GAP repeat after two conserved motifs in the repeat. The FG-GAP repeats are found in the N terminus of integrin alpha chains, a region that has been shown to be important for ligand binding. A putative Ca2+ binding motif is found in some of the repeats. Pssm-ID: 460357 Cd Length: 36 Bit Score: 50.97 E-value: 1.58e-08
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
56-114 | 3.74e-08 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 50.84 E-value: 3.74e-08
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
455-498 | 1.14e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 43.90 E-value: 1.14e-05
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
268-310 | 8.53e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 41.21 E-value: 8.53e-05
|
|||||||||||
| Name | Accession | Description | Interval | E-value | |||||||
| Integrin_alpha2 | pfam08441 | Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C ... |
490-921 | 0e+00 | |||||||
Integrin alpha; This domain is found in integrin alpha and integrin alpha precursors to the C terminus of a number of pfam01839 repeats and to the N-terminus of the pfam00357 cytoplasmic region. This region is composed of three immunoglobulin-like domains. Pssm-ID: 462478 [Multi-domain] Cd Length: 449 Bit Score: 560.40 E-value: 0e+00
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
389-443 | 3.43e-14 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 67.79 E-value: 3.43e-14
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
322-379 | 1.34e-12 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 63.16 E-value: 1.34e-12
|
|||||||||||
| FG-GAP | pfam01839 | FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven ... |
326-368 | 1.00e-09 | |||||||
FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven copies in alpha integrins. This repeat has been predicted to fold into a beta propeller structure. The repeat is called the FG-GAP repeat after two conserved motifs in the repeat. The FG-GAP repeats are found in the N terminus of integrin alpha chains, a region that has been shown to be important for ligand binding. A putative Ca2+ binding motif is found in some of the repeats. Pssm-ID: 460357 Cd Length: 36 Bit Score: 54.44 E-value: 1.00e-09
|
|||||||||||
| FG-GAP | pfam01839 | FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven ... |
392-428 | 1.58e-08 | |||||||
FG-GAP repeat; This family contains the extracellular repeat that is found in up to seven copies in alpha integrins. This repeat has been predicted to fold into a beta propeller structure. The repeat is called the FG-GAP repeat after two conserved motifs in the repeat. The FG-GAP repeats are found in the N terminus of integrin alpha chains, a region that has been shown to be important for ligand binding. A putative Ca2+ binding motif is found in some of the repeats. Pssm-ID: 460357 Cd Length: 36 Bit Score: 50.97 E-value: 1.58e-08
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
56-114 | 3.74e-08 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 50.84 E-value: 3.74e-08
|
|||||||||||
| FG-GAP_3 | pfam13517 | FG-GAP-like repeat; This entry represents a repeat found in alpha integrins and related ... |
334-412 | 2.59e-06 | |||||||
FG-GAP-like repeat; This entry represents a repeat found in alpha integrins and related proteins in which form a 7-fold repeat that adopts a beta-propeller fold. This repeat contains a putative calcium-binding site. These repeats are found in multiple proteins from eukaryotes and bacteria and mediate diverse biological processes at both molecular and cellular levels, such as cell-cell interactions, host-pathogen recognition or innate immune responses. Pssm-ID: 433275 [Multi-domain] Cd Length: 61 Bit Score: 45.68 E-value: 2.59e-06
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
455-498 | 1.14e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 43.90 E-value: 1.14e-05
|
|||||||||||
| Int_alpha | smart00191 | Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate ... |
268-310 | 8.53e-05 | |||||||
Integrin alpha (beta-propellor repeats); Integrins are cell adhesion molecules that mediate cell-extracellular matrix and cell-cell interactions. They contain both alpha and beta subunits. Alpha integrins are proposed to contain a domain containing a 7-fold repeat that adopts a beta-propellor fold. Some of these domains contain an inserted von Willebrand factor type-A domain. Some repeats contain putative calcium-binding sites. The 7-fold repeat domain is homologous to a similar domain in phosphatidylinositol-glycan-specific phospholipase D. Pssm-ID: 214549 [Multi-domain] Cd Length: 57 Bit Score: 41.21 E-value: 8.53e-05
|
|||||||||||
| FG-GAP_3 | pfam13517 | FG-GAP-like repeat; This entry represents a repeat found in alpha integrins and related ... |
401-476 | 1.54e-03 | |||||||
FG-GAP-like repeat; This entry represents a repeat found in alpha integrins and related proteins in which form a 7-fold repeat that adopts a beta-propeller fold. This repeat contains a putative calcium-binding site. These repeats are found in multiple proteins from eukaryotes and bacteria and mediate diverse biological processes at both molecular and cellular levels, such as cell-cell interactions, host-pathogen recognition or innate immune responses. Pssm-ID: 433275 [Multi-domain] Cd Length: 61 Bit Score: 37.98 E-value: 1.54e-03
|
|||||||||||
| Blast search parameters | ||||
|