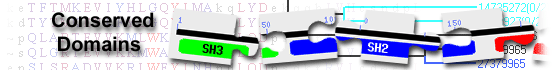growth arrest-specific protein 1 precursor [Homo sapiens]
GDNF domain-containing protein( domain architecture ID 10658247)
GDNF domain-containing protein similar to Homo sapiens growth arrest-specific protein 1 (GAS-1), which is a specific growth arrest protein involved in growth suppression
List of domain hits

| Name | Accession | Description | Interval | E-value | |||
| GDNF | smart00907 | GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 ... |
48-143 | 2.46e-04 | |||
GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 proteins. GDNF and neurturin (NTN) receptors are potent survival factors for sympathetic, sensory and central nervous system neurons.. GDNF and neurturin promote neuronal survival by signaling through similar multicomponent receptors that consist of a common receptor tyrosine kinase and a member of a GPI-linked family of receptors that determines ligand specificity. : Pssm-ID: 197975 Cd Length: 86 Bit Score: 39.55 E-value: 2.46e-04
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| GDNF | smart00907 | GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 ... |
48-143 | 2.46e-04 | |||
GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 proteins. GDNF and neurturin (NTN) receptors are potent survival factors for sympathetic, sensory and central nervous system neurons.. GDNF and neurturin promote neuronal survival by signaling through similar multicomponent receptors that consist of a common receptor tyrosine kinase and a member of a GPI-linked family of receptors that determines ligand specificity. Pssm-ID: 197975 Cd Length: 86 Bit Score: 39.55 E-value: 2.46e-04
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| GDNF | smart00907 | GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 ... |
48-143 | 2.46e-04 | |||
GDNF/GAS1 domain; This cysteine rich domain is found in multiple copies in GNDF and GAS1 proteins. GDNF and neurturin (NTN) receptors are potent survival factors for sympathetic, sensory and central nervous system neurons.. GDNF and neurturin promote neuronal survival by signaling through similar multicomponent receptors that consist of a common receptor tyrosine kinase and a member of a GPI-linked family of receptors that determines ligand specificity. Pssm-ID: 197975 Cd Length: 86 Bit Score: 39.55 E-value: 2.46e-04
|
|||||||
| Blast search parameters | ||||
|