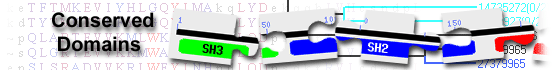cytochrome c oxidase assembly factor 8 isoform 1 [Homo sapiens]
APOPT family protein( domain architecture ID 10562973)
APOPT family protein is a DUF2315 domain-containing protein similar to Homo sapiens mitochondrial apoptogenic protein 1, which is required for cytochrome c complex (COX) IV assembly and function
List of domain hits

| Name | Accession | Description | Interval | E-value | |||
| COA8 | pfam10231 | Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome ... |
53-181 | 3.56e-69 | |||
Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome c oxidase assembly factor 8), previously thought to induce apoptosis through the Cyclophilin-D-dependent pathway, is required for cytochrome c oxidase (COX) assembly and function. Apop1 is controlled by the ubiquitination-proteasome system (UPS). In Drosophila melanogaster, Apop1 is also involved in neurological impairment, COX enzymatic activity, and resistance to oxidative stress conditions. : Pssm-ID: 463011 Cd Length: 119 Bit Score: 206.34 E-value: 3.56e-69
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| COA8 | pfam10231 | Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome ... |
53-181 | 3.56e-69 | |||
Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome c oxidase assembly factor 8), previously thought to induce apoptosis through the Cyclophilin-D-dependent pathway, is required for cytochrome c oxidase (COX) assembly and function. Apop1 is controlled by the ubiquitination-proteasome system (UPS). In Drosophila melanogaster, Apop1 is also involved in neurological impairment, COX enzymatic activity, and resistance to oxidative stress conditions. Pssm-ID: 463011 Cd Length: 119 Bit Score: 206.34 E-value: 3.56e-69
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| COA8 | pfam10231 | Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome ... |
53-181 | 3.56e-69 | |||
Cytochrome c oxidase assembly factor 8; Apoptogenic protein 1 (Apop1, also known as cytochrome c oxidase assembly factor 8), previously thought to induce apoptosis through the Cyclophilin-D-dependent pathway, is required for cytochrome c oxidase (COX) assembly and function. Apop1 is controlled by the ubiquitination-proteasome system (UPS). In Drosophila melanogaster, Apop1 is also involved in neurological impairment, COX enzymatic activity, and resistance to oxidative stress conditions. Pssm-ID: 463011 Cd Length: 119 Bit Score: 206.34 E-value: 3.56e-69
|
|||||||
| Blast search parameters | ||||
|