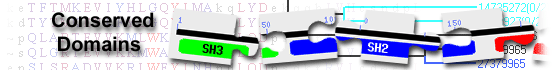diacylglycerol kinase alpha isoform 1 [Mus musculus]
List of domain hits

| Name | Accession | Description | Interval | E-value | ||||
| DAGKa | smart00045 | Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
515-696 | 1.23e-74 | ||||
Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain might either be an accessory domain or else contribute to the catalytic domain. Bacterial homologues are known. : Pssm-ID: 214486 Cd Length: 160 Bit Score: 238.00 E-value: 1.23e-74
|
||||||||
| DAGKc | smart00046 | Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
374-494 | 5.45e-52 | ||||
Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain is presumed to be the catalytic domain. Bacterial homologues areknown. : Pssm-ID: 214487 [Multi-domain] Cd Length: 124 Bit Score: 176.33 E-value: 5.45e-52
|
||||||||
| DAG_kinase_N super family | cl20554 | Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol ... |
4-111 | 8.01e-37 | ||||
Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol kinases. The actual alignment was detected with superfamily member pfam14513: Pssm-ID: 464196 Cd Length: 135 Bit Score: 134.43 E-value: 8.01e-37
|
||||||||
| C1_DGKalpha_rpt2 | cd20890 | second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha ... |
271-332 | 1.40e-34 | ||||
second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha (DAG kinase alpha) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase alpha, also called 80 kDa diacylglycerol kinase, or diglyceride kinase alpha (DGK-alpha), converts the second messenger diacylglycerol into phosphatidate upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DAG kinase alpha contains two copies of the C1 domain. This model corresponds to the second one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. : Pssm-ID: 410440 Cd Length: 62 Bit Score: 125.73 E-value: 1.40e-34
|
||||||||
| C1 super family | cl00040 | protein kinase C conserved region 1 (C1 domain) superfamily; The C1 domain is a cysteine-rich ... |
202-262 | 3.31e-24 | ||||
protein kinase C conserved region 1 (C1 domain) superfamily; The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains. It contains the motif HX12CX2CXnCX2CX4HX2CX7C, where C and H are cysteine and histidine, respectively; X represents other residues; and n is either 13 or 14. C1 has a globular fold with two separate Zn(2+)-binding sites. It was originally discovered as lipid-binding modules in protein kinase C (PKC) isoforms. C1 domains that bind and respond to phorbol esters (PE) and diacylglycerol (DAG) are referred to as typical, and those that do not respond to PE and DAG are deemed atypical. A C1 domain may also be referred to as PKC or non-PKC C1, based on the parent protein's activity. Most C1 domain-containing non-PKC proteins act as lipid kinases and scaffolds, except PKD which acts as a protein kinase. PKC C1 domains play roles in membrane translocation and activation of the enzyme. The actual alignment was detected with superfamily member cd20799: Pssm-ID: 412127 Cd Length: 62 Bit Score: 95.90 E-value: 3.31e-24
|
||||||||
| EFh | cd00051 | EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal ... |
115-184 | 8.01e-13 | ||||
EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal modulators; most examples in this alignment model have 2 active canonical EF hands. Ca2+ binding induces a conformational change in the EF-hand motif, leading to the activation or inactivation of target proteins. EF-hands tend to occur in pairs or higher copy numbers. : Pssm-ID: 238008 [Multi-domain] Cd Length: 63 Bit Score: 63.72 E-value: 8.01e-13
|
||||||||
| Name | Accession | Description | Interval | E-value | ||||||
| DAGKa | smart00045 | Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
515-696 | 1.23e-74 | ||||||
Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain might either be an accessory domain or else contribute to the catalytic domain. Bacterial homologues are known. Pssm-ID: 214486 Cd Length: 160 Bit Score: 238.00 E-value: 1.23e-74
|
||||||||||
| DAGK_acc | pfam00609 | Diacylglycerol kinase accessory domain; Diacylglycerol (DAG) is a second messenger that acts ... |
515-696 | 5.96e-63 | ||||||
Diacylglycerol kinase accessory domain; Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. This domain is assumed to be an accessory domain: its function is unknown. Pssm-ID: 459866 Cd Length: 158 Bit Score: 207.07 E-value: 5.96e-63
|
||||||||||
| DAGKc | smart00046 | Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
374-494 | 5.45e-52 | ||||||
Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain is presumed to be the catalytic domain. Bacterial homologues areknown. Pssm-ID: 214487 [Multi-domain] Cd Length: 124 Bit Score: 176.33 E-value: 5.45e-52
|
||||||||||
| DAG_kinase_N | pfam14513 | Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol ... |
4-111 | 8.01e-37 | ||||||
Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol kinases. Pssm-ID: 464196 Cd Length: 135 Bit Score: 134.43 E-value: 8.01e-37
|
||||||||||
| C1_DGKalpha_rpt2 | cd20890 | second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha ... |
271-332 | 1.40e-34 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha (DAG kinase alpha) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase alpha, also called 80 kDa diacylglycerol kinase, or diglyceride kinase alpha (DGK-alpha), converts the second messenger diacylglycerol into phosphatidate upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DAG kinase alpha contains two copies of the C1 domain. This model corresponds to the second one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410440 Cd Length: 62 Bit Score: 125.73 E-value: 1.40e-34
|
||||||||||
| DAGK_cat | pfam00781 | Diacylglycerol kinase catalytic domain; Diacylglycerol (DAG) is a second messenger that acts ... |
372-489 | 2.31e-33 | ||||||
Diacylglycerol kinase catalytic domain; Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. The catalytic domain is assumed from the finding of bacterial homologs. YegS is the Escherichia coli protein in this family whose crystal structure reveals an active site in the inter-domain cleft formed by four conserved sequence motifs, revealing a novel metal-binding site. The residues of this site are conserved across the family. Pssm-ID: 425868 [Multi-domain] Cd Length: 125 Bit Score: 124.23 E-value: 2.31e-33
|
||||||||||
| C1_DGK_typeI_rpt1 | cd20799 | first protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; ... |
202-262 | 3.31e-24 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. Type I DAG kinases (DGKs) contain EF-hand structures that bind Ca(2+) and recoverin homology domains, in addition to C1 and catalytic domains that are present in all DGKs. Type I DGKs, regulated by calcium binding, include three DGK isozymes (alpha, beta and gamma). DAG kinase alpha, also called 80 kDa DAG kinase, or diglyceride kinase alpha (DGK-alpha), is active upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. DAG kinase beta, also called 90 kDa DAG kinase, or diglyceride kinase beta (DGK-beta), exhibits high phosphorylation activity for long-chain diacylglycerols. DAG kinase gamma, also called diglyceride kinase gamma (DGK-gamma), reverses the normal flow of glycerolipid biosynthesis by phosphorylating diacylglycerol back to phosphatidic acid. Members of this family contain two copies of the C1 domain. This model corresponds to the first one. DGK-alpha contains atypical C1 domains, while DGK-beta and DGK-gamma contain typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410349 Cd Length: 62 Bit Score: 95.90 E-value: 3.31e-24
|
||||||||||
| LCB5 | COG1597 | Phosphatidylglycerol kinase, diacylglycerol kinase family [Lipid transport and metabolism, ... |
373-709 | 9.10e-16 | ||||||
Phosphatidylglycerol kinase, diacylglycerol kinase family [Lipid transport and metabolism, General function prediction only]; Pssm-ID: 441205 [Multi-domain] Cd Length: 295 Bit Score: 78.36 E-value: 9.10e-16
|
||||||||||
| EFh | cd00051 | EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal ... |
115-184 | 8.01e-13 | ||||||
EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal modulators; most examples in this alignment model have 2 active canonical EF hands. Ca2+ binding induces a conformational change in the EF-hand motif, leading to the activation or inactivation of target proteins. EF-hands tend to occur in pairs or higher copy numbers. Pssm-ID: 238008 [Multi-domain] Cd Length: 63 Bit Score: 63.72 E-value: 8.01e-13
|
||||||||||
| C1_1 | pfam00130 | Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the ... |
271-323 | 2.07e-11 | ||||||
Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the Protein kinase C conserved region 1 (C1) domain. Pssm-ID: 395079 Cd Length: 53 Bit Score: 59.38 E-value: 2.07e-11
|
||||||||||
| C1_1 | pfam00130 | Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the ... |
207-255 | 1.41e-10 | ||||||
Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the Protein kinase C conserved region 1 (C1) domain. Pssm-ID: 395079 Cd Length: 53 Bit Score: 57.07 E-value: 1.41e-10
|
||||||||||
| C1 | smart00109 | Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol ... |
207-254 | 1.20e-08 | ||||||
Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol esters and diacylglycerol. Some bind RasGTP. Zinc-binding domains. Pssm-ID: 197519 Cd Length: 50 Bit Score: 51.31 E-value: 1.20e-08
|
||||||||||
| EF-hand_7 | pfam13499 | EF-hand domain pair; |
113-180 | 4.40e-07 | ||||||
EF-hand domain pair; Pssm-ID: 463900 [Multi-domain] Cd Length: 67 Bit Score: 47.63 E-value: 4.40e-07
|
||||||||||
| FRQ1 | COG5126 | Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; |
113-184 | 7.61e-07 | ||||||
Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; Pssm-ID: 444056 [Multi-domain] Cd Length: 137 Bit Score: 49.02 E-value: 7.61e-07
|
||||||||||
| PRK13059 | PRK13059 | putative lipid kinase; Reviewed |
424-499 | 3.15e-05 | ||||||
putative lipid kinase; Reviewed Pssm-ID: 183858 Cd Length: 295 Bit Score: 46.57 E-value: 3.15e-05
|
||||||||||
| PTZ00184 | PTZ00184 | calmodulin; Provisional |
113-183 | 7.99e-05 | ||||||
calmodulin; Provisional Pssm-ID: 185504 [Multi-domain] Cd Length: 149 Bit Score: 43.60 E-value: 7.99e-05
|
||||||||||
| EFh | smart00054 | EF-hand, calcium binding motif; EF-hands are calcium-binding motifs that occur at least in ... |
161-184 | 2.57e-04 | ||||||
EF-hand, calcium binding motif; EF-hands are calcium-binding motifs that occur at least in pairs. Links between disease states and genes encoding EF-hands, particularly the S100 subclass, are emerging. Each motif consists of a 12 residue loop flanked on either side by a 12 residue alpha-helix. EF-hands undergo a conformational change unpon binding calcium ions. Pssm-ID: 197492 [Multi-domain] Cd Length: 29 Bit Score: 38.51 E-value: 2.57e-04
|
||||||||||
| C1 | smart00109 | Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol ... |
271-320 | 2.60e-04 | ||||||
Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol esters and diacylglycerol. Some bind RasGTP. Zinc-binding domains. Pssm-ID: 197519 Cd Length: 50 Bit Score: 39.37 E-value: 2.60e-04
|
||||||||||
| Name | Accession | Description | Interval | E-value | ||||||
| DAGKa | smart00045 | Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
515-696 | 1.23e-74 | ||||||
Diacylglycerol kinase accessory domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain might either be an accessory domain or else contribute to the catalytic domain. Bacterial homologues are known. Pssm-ID: 214486 Cd Length: 160 Bit Score: 238.00 E-value: 1.23e-74
|
||||||||||
| DAGK_acc | pfam00609 | Diacylglycerol kinase accessory domain; Diacylglycerol (DAG) is a second messenger that acts ... |
515-696 | 5.96e-63 | ||||||
Diacylglycerol kinase accessory domain; Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. This domain is assumed to be an accessory domain: its function is unknown. Pssm-ID: 459866 Cd Length: 158 Bit Score: 207.07 E-value: 5.96e-63
|
||||||||||
| DAGKc | smart00046 | Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger ... |
374-494 | 5.45e-52 | ||||||
Diacylglycerol kinase catalytic domain (presumed); Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. DAG can be produced from the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by a phosphoinositide-specific phospholipase C and by the degradation of phosphatidylcholine (PC) by a phospholipase C or the concerted actions of phospholipase D and phosphatidate phosphohydrolase. This domain is presumed to be the catalytic domain. Bacterial homologues areknown. Pssm-ID: 214487 [Multi-domain] Cd Length: 124 Bit Score: 176.33 E-value: 5.45e-52
|
||||||||||
| DAG_kinase_N | pfam14513 | Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol ... |
4-111 | 8.01e-37 | ||||||
Diacylglycerol kinase N-terminus; This domain is found at the N-terminus of diacylglycerol kinases. Pssm-ID: 464196 Cd Length: 135 Bit Score: 134.43 E-value: 8.01e-37
|
||||||||||
| C1_DGKalpha_rpt2 | cd20890 | second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha ... |
271-332 | 1.40e-34 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase alpha (DAG kinase alpha) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase alpha, also called 80 kDa diacylglycerol kinase, or diglyceride kinase alpha (DGK-alpha), converts the second messenger diacylglycerol into phosphatidate upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DAG kinase alpha contains two copies of the C1 domain. This model corresponds to the second one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410440 Cd Length: 62 Bit Score: 125.73 E-value: 1.40e-34
|
||||||||||
| DAGK_cat | pfam00781 | Diacylglycerol kinase catalytic domain; Diacylglycerol (DAG) is a second messenger that acts ... |
372-489 | 2.31e-33 | ||||||
Diacylglycerol kinase catalytic domain; Diacylglycerol (DAG) is a second messenger that acts as a protein kinase C activator. The catalytic domain is assumed from the finding of bacterial homologs. YegS is the Escherichia coli protein in this family whose crystal structure reveals an active site in the inter-domain cleft formed by four conserved sequence motifs, revealing a novel metal-binding site. The residues of this site are conserved across the family. Pssm-ID: 425868 [Multi-domain] Cd Length: 125 Bit Score: 124.23 E-value: 2.31e-33
|
||||||||||
| C1_DGK_typeI_like_rpt2 | cd20851 | second protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; ... |
271-323 | 2.40e-25 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. Type I DAG kinases (DGKs) contain EF-hand structures that bind Ca(2+) and recoverin homology domains, in addition to C1 and catalytic domains that are present in all DGKs. Type I DGKs, regulated by calcium binding, include three DGK isozymes (alpha, beta and gamma). DAG kinase alpha, also called 80 kDa DAG kinase, or diglyceride kinase alpha (DGK-alpha), is active upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. DAG kinase beta, also called 90 kDa DAG kinase, or diglyceride kinase beta (DGK-beta), exhibits high phosphorylation activity for long-chain diacylglycerols. DAG kinase gamma, also called diglyceride kinase gamma (DGK-gamma), reverses the normal flow of glycerolipid biosynthesis by phosphorylating diacylglycerol back to phosphatidic acid. Members of this family contain two copies of the C1 domain. This model corresponds to the second one. DGK-alpha contains atypical C1 domains, while DGK-beta and DGK-gamma contain typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410401 Cd Length: 52 Bit Score: 98.96 E-value: 2.40e-25
|
||||||||||
| C1_DGK_typeI_rpt1 | cd20799 | first protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; ... |
202-262 | 3.31e-24 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in type I diacylglycerol kinases; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. Type I DAG kinases (DGKs) contain EF-hand structures that bind Ca(2+) and recoverin homology domains, in addition to C1 and catalytic domains that are present in all DGKs. Type I DGKs, regulated by calcium binding, include three DGK isozymes (alpha, beta and gamma). DAG kinase alpha, also called 80 kDa DAG kinase, or diglyceride kinase alpha (DGK-alpha), is active upon cell stimulation, initiating the resynthesis of phosphatidylinositols and attenuating protein kinase C activity. DAG kinase beta, also called 90 kDa DAG kinase, or diglyceride kinase beta (DGK-beta), exhibits high phosphorylation activity for long-chain diacylglycerols. DAG kinase gamma, also called diglyceride kinase gamma (DGK-gamma), reverses the normal flow of glycerolipid biosynthesis by phosphorylating diacylglycerol back to phosphatidic acid. Members of this family contain two copies of the C1 domain. This model corresponds to the first one. DGK-alpha contains atypical C1 domains, while DGK-beta and DGK-gamma contain typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410349 Cd Length: 62 Bit Score: 95.90 E-value: 3.31e-24
|
||||||||||
| C1_DGKbeta_rpt1 | cd20845 | first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase beta (DAG ... |
200-264 | 1.78e-21 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase beta (DAG kinase beta) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase beta, also called 90 kDa diacylglycerol kinase, or diglyceride kinase beta (DGK-beta), exhibits high phosphorylation activity for long-chain diacylglycerols. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DAG kinase beta contains two copies of the C1 domain. This model corresponds to the first one. DGK-beta contains typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410395 Cd Length: 66 Bit Score: 88.37 E-value: 1.78e-21
|
||||||||||
| C1_DGKgamma_rpt1 | cd20846 | first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase gamma ... |
191-262 | 1.64e-20 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase gamma (DAG kinase gamma) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase gamma, also called diglyceride kinase gamma (DGK-gamma), reverses the normal flow of glycerolipid biosynthesis by phosphorylating diacylglycerol back to phosphatidic acid. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DGK-gamma contains two copies of the C1 domain. This model corresponds to the first one. DGK-gamma contains typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410396 Cd Length: 73 Bit Score: 85.75 E-value: 1.64e-20
|
||||||||||
| C1_DGKbeta_rpt2 | cd20891 | second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase beta ... |
271-328 | 2.29e-20 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase beta (DAG kinase beta) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase beta, also called 90 kDa diacylglycerol kinase, or diglyceride kinase beta (DGK-beta), exhibits high phosphorylation activity for long-chain diacylglycerols. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DAG kinase beta contains two copies of the C1 domain. This model corresponds to the second one. DGK-beta contains typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410441 Cd Length: 59 Bit Score: 85.04 E-value: 2.29e-20
|
||||||||||
| C1_DGKgamma_rpt2 | cd20892 | second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase gamma ... |
271-332 | 9.47e-20 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase gamma (DAG kinase gamma) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase gamma, also called diglyceride kinase gamma (DGK-gamma), reverses the normal flow of glycerolipid biosynthesis by phosphorylating diacylglycerol back to phosphatidic acid. It is classified as a type I DAG kinase (DGK), containing EF-hand structures that bind Ca(2+) and a recoverin homology domain, in addition to C1 and catalytic domains that are present in all DGKs. As a type I DGK, it is regulated by calcium binding. DGK-gamma contains two copies of the C1 domain. This model corresponds to the second one. DGK-gamma contains typical C1 domains that bind DAG and phorbol esters. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410442 Cd Length: 61 Bit Score: 83.32 E-value: 9.47e-20
|
||||||||||
| LCB5 | COG1597 | Phosphatidylglycerol kinase, diacylglycerol kinase family [Lipid transport and metabolism, ... |
373-709 | 9.10e-16 | ||||||
Phosphatidylglycerol kinase, diacylglycerol kinase family [Lipid transport and metabolism, General function prediction only]; Pssm-ID: 441205 [Multi-domain] Cd Length: 295 Bit Score: 78.36 E-value: 9.10e-16
|
||||||||||
| C1_DGK_rpt2 | cd20805 | second protein kinase C conserved region 1 (C1 domain) found in the diacylglycerol kinase ... |
271-323 | 1.20e-14 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in the diacylglycerol kinase family; The diacylglycerol kinase (DGK, EC 2.7.1.107) family of enzymes plays critical roles in lipid signaling pathways by converting diacylglycerol to phosphatidic acid, thereby downregulating signaling by the former and upregulating signaling by the latter second messenger. Ten DGK family isozymes have been identified to date, which possess different interaction motifs imparting distinct temporal and spatial control of DGK activity to each isozyme. They have been classified into five types (I-V), according to domain architecture and some common features. All DGK isozymes, except for DGKtheta, contain two copies of the C1 domain. This model corresponds to the second one. DGKtheta harbors three C1 domains. Its third C1 domain is included here. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410355 Cd Length: 55 Bit Score: 68.63 E-value: 1.20e-14
|
||||||||||
| EFh | cd00051 | EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal ... |
115-184 | 8.01e-13 | ||||||
EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal modulators; most examples in this alignment model have 2 active canonical EF hands. Ca2+ binding induces a conformational change in the EF-hand motif, leading to the activation or inactivation of target proteins. EF-hands tend to occur in pairs or higher copy numbers. Pssm-ID: 238008 [Multi-domain] Cd Length: 63 Bit Score: 63.72 E-value: 8.01e-13
|
||||||||||
| C1_1 | pfam00130 | Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the ... |
271-323 | 2.07e-11 | ||||||
Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the Protein kinase C conserved region 1 (C1) domain. Pssm-ID: 395079 Cd Length: 53 Bit Score: 59.38 E-value: 2.07e-11
|
||||||||||
| C1_1 | pfam00130 | Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the ... |
207-255 | 1.41e-10 | ||||||
Phorbol esters/diacylglycerol binding domain (C1 domain); This domain is also known as the Protein kinase C conserved region 1 (C1) domain. Pssm-ID: 395079 Cd Length: 53 Bit Score: 57.07 E-value: 1.41e-10
|
||||||||||
| C1_DGK_typeII_rpt1 | cd20800 | first protein kinase C conserved region 1 (C1 domain) found in type II diacylglycerol kinases; ... |
203-260 | 3.77e-10 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in type II diacylglycerol kinases; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. Type II DAG kinases (DGKs) contain pleckstrin homology (PH) and sterile alpha motifs (SAM) domains, in addition to C1 and catalytic domains that are present in all DGKs. The SAM domain mediates oligomerization of type II DGKs. Three DGK isozymes (delta, eta and kappa) are classified as type II. DAG kinase delta, also called 130 kDa DAG kinase, or diglyceride kinase delta (DGK-delta), is a residential lipid kinase in the endoplasmic reticulum. It promotes lipogenesis and is involved in triglyceride biosynthesis. DAG kinase eta, also called diglyceride kinase eta (DGK-eta), plays a key role in promoting cell growth. The DAG kinase eta gene, DGKH, is a replicated risk gene of bipolar disorder (BPD). DAG kinase kappa is also called diglyceride kinase kappa (DGK-kappa) or 142 kDa DAG kinase. Members of this family contain two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410350 Cd Length: 60 Bit Score: 56.18 E-value: 3.77e-10
|
||||||||||
| C1 | cd00029 | protein kinase C conserved region 1 (C1 domain) superfamily; The C1 domain is a cysteine-rich ... |
207-254 | 6.79e-09 | ||||||
protein kinase C conserved region 1 (C1 domain) superfamily; The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains. It contains the motif HX12CX2CXnCX2CX4HX2CX7C, where C and H are cysteine and histidine, respectively; X represents other residues; and n is either 13 or 14. C1 has a globular fold with two separate Zn(2+)-binding sites. It was originally discovered as lipid-binding modules in protein kinase C (PKC) isoforms. C1 domains that bind and respond to phorbol esters (PE) and diacylglycerol (DAG) are referred to as typical, and those that do not respond to PE and DAG are deemed atypical. A C1 domain may also be referred to as PKC or non-PKC C1, based on the parent protein's activity. Most C1 domain-containing non-PKC proteins act as lipid kinases and scaffolds, except PKD which acts as a protein kinase. PKC C1 domains play roles in membrane translocation and activation of the enzyme. Pssm-ID: 410341 Cd Length: 50 Bit Score: 52.13 E-value: 6.79e-09
|
||||||||||
| C1 | smart00109 | Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol ... |
207-254 | 1.20e-08 | ||||||
Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol esters and diacylglycerol. Some bind RasGTP. Zinc-binding domains. Pssm-ID: 197519 Cd Length: 50 Bit Score: 51.31 E-value: 1.20e-08
|
||||||||||
| C1_DGKtheta_typeV_rpt3 | cd20854 | third protein kinase C conserved region 1 (C1 domain) found in type V diacylglycerol kinase, ... |
271-332 | 2.10e-08 | ||||||
third protein kinase C conserved region 1 (C1 domain) found in type V diacylglycerol kinase, DAG kinase theta, and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase theta, also called diglyceride kinase theta (DGK-theta), is the only isoform classified as type V; it contains a pleckstrin homology (PH)-like domain and an additional C1 domain, compared to other DGKs. It may regulate the activity of protein kinase C by controlling the balance between the two signaling lipids, diacylglycerol and phosphatidic acid. DAG kinase theta contains three copies of the C1 domain. This model corresponds to the third one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410404 Cd Length: 63 Bit Score: 51.11 E-value: 2.10e-08
|
||||||||||
| C1_DGKepsilon_typeIII_rpt2 | cd20853 | second protein kinase C conserved region 1 (C1 domain) found in type III diacylglycerol kinase, ... |
271-332 | 7.16e-08 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in type III diacylglycerol kinase, DAG kinase epsilon, and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase epsilon, also called diglyceride kinase epsilon (DGK-epsilon), is the only isoform classified as type III; it possesses a hydrophobic domain in addition to C1 and catalytic domains that are present in all DGKs, and shows selectivity for acyl chains. It is highly selective for arachidonate-containing species of DAG. It may terminate signals transmitted through arachidonoyl-DAG or may contribute to the synthesis of phospholipids with defined fatty acid composition. DAG kinase epsilon contains two copies of the C1 domain. This model corresponds to the second one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410403 Cd Length: 63 Bit Score: 49.58 E-value: 7.16e-08
|
||||||||||
| EF-hand_7 | pfam13499 | EF-hand domain pair; |
113-180 | 4.40e-07 | ||||||
EF-hand domain pair; Pssm-ID: 463900 [Multi-domain] Cd Length: 67 Bit Score: 47.63 E-value: 4.40e-07
|
||||||||||
| FRQ1 | COG5126 | Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; |
113-184 | 7.61e-07 | ||||||
Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; Pssm-ID: 444056 [Multi-domain] Cd Length: 137 Bit Score: 49.02 E-value: 7.61e-07
|
||||||||||
| C1_SpBZZ1-like | cd20824 | protein kinase C conserved region 1 (C1 domain) found in Schizosaccharomyces pombe protein ... |
207-253 | 8.66e-07 | ||||||
protein kinase C conserved region 1 (C1 domain) found in Schizosaccharomyces pombe protein BZZ1 and similar proteins; BZZ1 is a syndapin-like F-BAR protein that plays a role in endocytosis and trafficking to the vacuole. It functions with type I myosins to restore polarity of the actin cytoskeleton after NaCl stress. BZZ1 contains an N-terminal F-BAR (FES-CIP4 Homology and Bin/Amphiphysin/Rvs), a central coiled-coil, and two C-terminal SH3 domains. Schizosaccharomyces pombe BZZ1 also harbors a C1 domain, but Saccharomyces cerevisiae BZZ1 doesn't have any. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410374 Cd Length: 53 Bit Score: 46.16 E-value: 8.66e-07
|
||||||||||
| FRQ1 | COG5126 | Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; |
112-184 | 2.09e-06 | ||||||
Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]; Pssm-ID: 444056 [Multi-domain] Cd Length: 137 Bit Score: 47.86 E-value: 2.09e-06
|
||||||||||
| C1_aPKC | cd20794 | protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) ... |
207-247 | 4.76e-06 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) family; PKCs are classified into three groups (classical, atypical, and novel) depending on their mode of activation and the structural characteristics of their regulatory domain. aPKCs only require phosphatidylserine (PS) for activation. They contain a C2-like region, instead of a calcium-binding (C2) region found in classical PKCs, in their regulatory domain. There are two aPKC isoforms, zeta and iota. aPKCs are involved in many cellular functions including proliferation, migration, apoptosis, polarity maintenance and cytoskeletal regulation. They also play a critical role in the regulation of glucose metabolism and in the pathogenesis of type 2 diabetes. PKC-zeta plays a critical role in activating the glucose transport response. It is activated by glucose, insulin, and exercise through diverse pathways. PKC-zeta also plays a central role in maintaining cell polarity in yeast and mammalian cells. In addition, it affects actin remodeling in muscle cells. PKC-iota is directly implicated in carcinogenesis. It is critical to oncogenic signaling mediated by Ras and Bcr-Abl. The PKC-iota gene is the target of tumor-specific gene amplification in many human cancers, and has been identified as a human oncogene. In addition to its role in transformed growth, PKC-iota also promotes invasion, chemoresistance, and tumor cell survival. Expression profiling of PKC-iota is a prognostic marker of poor clinical outcome in several human cancers. PKC-iota also plays a role in establishing cell polarity, and has critical embryonic functions. Members of this family contain one C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410344 Cd Length: 55 Bit Score: 44.18 E-value: 4.76e-06
|
||||||||||
| C1_DGKdelta_rpt1 | cd20847 | first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase delta ... |
205-265 | 4.93e-06 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase delta (DAG kinase delta) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase delta, also called 130 kDa diacylglycerol kinase, or diglyceride kinase delta (DGK-delta), is a residential lipid kinase in the endoplasmic reticulum. It promotes lipogenesis and is involved in triglyceride biosynthesis. It is classified as a type II DAG kinase (DGK), containing pleckstrin homology (PH) and sterile alpha motifs (SAM) domains, in addition to C1 and catalytic domains that are present in all DGKs. The SAM domain mediates oligomerization of type II DGKs. DAG kinase delta contains two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410397 Cd Length: 85 Bit Score: 45.09 E-value: 4.93e-06
|
||||||||||
| C1_dGM13116p-like | cd20831 | protein kinase C conserved region 1 (C1 domain) found in Drosophila melanogaster GM13116p and ... |
206-254 | 5.87e-06 | ||||||
protein kinase C conserved region 1 (C1 domain) found in Drosophila melanogaster GM13116p and similar proteins; This group contains uncharacterized proteins including Drosophila melanogaster GM13116p and Caenorhabditis elegans hypothetical protein R11G1.4, both of which contain C2 (a calcium-binding domain) and C1 domains. This model describes the C1 domain, a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410381 Cd Length: 58 Bit Score: 44.25 E-value: 5.87e-06
|
||||||||||
| C1_DGKeta_rpt1 | cd20848 | first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase eta (DAG ... |
205-260 | 1.01e-05 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in diacylglycerol kinase eta (DAG kinase eta) and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase eta, also called diglyceride kinase eta (DGK-eta), plays a key role in promoting cell growth. It is classified as a type II DAG kinase (DGK), containing pleckstrin homology (PH) and sterile alpha motifs (SAM) domains, in addition to C1 and catalytic domains that are present in all DGKs. The SAM domain mediates oligomerization of type II DGKs. The diacylglycerol kinase eta gene, DGKH, is a replicated risk gene of bipolar disorder (BPD). DAG kinase eta contains two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410398 Cd Length: 86 Bit Score: 44.38 E-value: 1.01e-05
|
||||||||||
| C1_aPKC_zeta | cd21095 | protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) ... |
207-247 | 1.41e-05 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) zeta type; PKCs are classified into three groups (classical, atypical, and novel) depending on their mode of activation and the structural characteristics of their regulatory domain. aPKCs only require phosphatidylserine (PS) for activation. They contain a C2-like region, instead of a calcium-binding (C2) region found in classical PKCs, in their regulatory domain. There are two aPKC isoforms, zeta and iota. aPKCs are involved in many cellular functions including proliferation, migration, apoptosis, polarity maintenance and cytoskeletal regulation. They also play a critical role in the regulation of glucose metabolism and in the pathogenesis of type 2 diabetes. PKC-zeta plays a critical role in activating the glucose transport response. It is activated by glucose, insulin, and exercise through diverse pathways. PKC-zeta also plays a central role in maintaining cell polarity in yeast and mammalian cells. In addition, it affects actin remodeling in muscle cells. Members of this family contain C1 domain found in aPKC isoform zeta. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410448 Cd Length: 55 Bit Score: 43.05 E-value: 1.41e-05
|
||||||||||
| PRK13059 | PRK13059 | putative lipid kinase; Reviewed |
424-499 | 3.15e-05 | ||||||
putative lipid kinase; Reviewed Pssm-ID: 183858 Cd Length: 295 Bit Score: 46.57 E-value: 3.15e-05
|
||||||||||
| C1_TNS2-like | cd20826 | protein kinase C conserved region 1 (C1 domain) found in tensin-2 like (TNS2-like) proteins; ... |
207-257 | 3.66e-05 | ||||||
protein kinase C conserved region 1 (C1 domain) found in tensin-2 like (TNS2-like) proteins; The TNS2-like group includes TNS2, and variants of TNS1 and TNS3. Tensin-2 (TNS2), also called C1 domain-containing phosphatase and tensin (C1-TEN), or tensin-like C1 domain-containing phosphatase (TENC1), is an essential component for the maintenance of glomerular basement membrane (GBM) structures. It regulates cell motility and proliferation. It may have phosphatase activity. TNS2 reduces AKT1 phosphorylation, lowers AKT1 kinase activity and interferes with AKT1 signaling. Tensin-1 (TNS1) plays a role in fibrillar adhesion formation. It may be involved in cell migration, cartilage development and in linking signal transduction pathways to the cytoskeleton. Tensin-3 (TNS3), also called tensin-like SH2 domain-containing protein 1 (TENS1), or tumor endothelial marker 6 (TEM6), may play a role in actin remodeling. It is involved in the dissociation of the integrin-tensin-actin complex. Typical TNS1 and TNS3 do not contain C1 domains, but some isoforms/variants do. Members of this family contain an N-terminal region with a zinc finger (C1 domain), a protein tyrosine phosphatase (PTP)-like domain and a protein kinase 2 (C2) domain, and a C-terminal region with SH2 and pTyr binding (PTB) domains. This model corresponds to C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410376 Cd Length: 52 Bit Score: 41.60 E-value: 3.66e-05
|
||||||||||
| PRK13054 | PRK13054 | lipid kinase; Reviewed |
423-464 | 6.68e-05 | ||||||
lipid kinase; Reviewed Pssm-ID: 237281 [Multi-domain] Cd Length: 300 Bit Score: 45.63 E-value: 6.68e-05
|
||||||||||
| PRK13055 | PRK13055 | putative lipid kinase; Reviewed |
424-483 | 6.75e-05 | ||||||
putative lipid kinase; Reviewed Pssm-ID: 237282 [Multi-domain] Cd Length: 334 Bit Score: 45.75 E-value: 6.75e-05
|
||||||||||
| PTZ00184 | PTZ00184 | calmodulin; Provisional |
113-183 | 7.99e-05 | ||||||
calmodulin; Provisional Pssm-ID: 185504 [Multi-domain] Cd Length: 149 Bit Score: 43.60 E-value: 7.99e-05
|
||||||||||
| C1_aPKC_iota | cd21094 | protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) ... |
207-247 | 8.53e-05 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the atypical protein kinase C (aPKC) iota type; PKCs are classified into three groups (classical, atypical, and novel) depending on their mode of activation and the structural characteristics of their regulatory domain. aPKCs only require phosphatidylserine (PS) for activation. They contain a C2-like region, instead of a calcium-binding (C2) region found in classical PKCs, in their regulatory domain. There are two aPKC isoforms, zeta and iota. aPKCs are involved in many cellular functions including proliferation, migration, apoptosis, polarity maintenance and cytoskeletal regulation. They also play a critical role in the regulation of glucose metabolism and in the pathogenesis of type 2 diabetes. PKC-iota is directly implicated in carcinogenesis. It is critical to oncogenic signaling mediated by Ras and Bcr-Abl. The PKC-iota gene is the target of tumor-specific gene amplification in many human cancers, and has been identified as a human oncogene. In addition to its role in transformed growth, PKC-iota also promotes invasion, chemoresistance, and tumor cell survival. Expression profiling of PKC-iota is a prognostic marker of poor clinical outcome in several human cancers. PKC-iota also plays a role in establishing cell polarity, and has critical embryonic functions. Members of this family contain C1 domain found in aPKC isoform iota. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410447 Cd Length: 55 Bit Score: 40.76 E-value: 8.53e-05
|
||||||||||
| C1_MgcRacGAP | cd20821 | protein kinase C conserved region 1 (C1 domain) found in male germ cell RacGap (MgcRacGAP) and ... |
207-254 | 1.27e-04 | ||||||
protein kinase C conserved region 1 (C1 domain) found in male germ cell RacGap (MgcRacGAP) and similar proteins; MgcRacGAP, also called Rac GTPase-activating protein 1 (RACGAP1) or protein CYK4, plays an important dual role in cytokinesis: i) it is part of centralspindlin-complex, together with the mitotic kinesin MKLP1, which is critical for the structure of the central spindle by promoting microtuble bundling; and ii) after phosphorylation by aurora B, MgcRacGAP becomes an effective regulator of RhoA and plays an important role in the assembly of the contractile ring and the initiation of cytokinesis. MgcRacGAP-like proteins contain an N-terminal C1 domain, and a C-terminal RhoGAP domain. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410371 Cd Length: 55 Bit Score: 40.08 E-value: 1.27e-04
|
||||||||||
| C1_TNS3_v | cd20889 | protein kinase C conserved region 1 (C1 domain) found in tensin-3 (TNS3) variant and similar ... |
207-254 | 1.58e-04 | ||||||
protein kinase C conserved region 1 (C1 domain) found in tensin-3 (TNS3) variant and similar proteins; Tensin-3 (TNS3), also called tensin-like SH2 domain-containing protein 1 (TENS1), or tumor endothelial marker 6 (TEM6), may play a role in actin remodeling. It is involved in the dissociation of the integrin-tensin-actin complex. This model corresponds to the C1 domain found in TNS3 variant. Typical TNS3 does not contain C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410439 Cd Length: 56 Bit Score: 39.87 E-value: 1.58e-04
|
||||||||||
| C1_MRCKgamma | cd20866 | protein kinase C conserved region 1 (C1 domain) found in myotonic dystrophy kinase-related ... |
207-254 | 1.98e-04 | ||||||
protein kinase C conserved region 1 (C1 domain) found in myotonic dystrophy kinase-related Cdc42-binding kinase gamma (MRCK gamma) and similar proteins; MRCK gamma (MRCKG), also called Cdc42-binding protein kinase gamma, DMPK-like gamma, myotonic dystrophy protein kinase-like gamma, or myotonic dystrophy protein kinase-like alpha, is a serine/threonine-protein kinase expressed in heart and skeletal muscles. It may act as a downstream effector of Cdc42 in cytoskeletal reorganization and contributes to the actomyosin contractility required for cell invasion, through the regulation of MYPT1 and thus MLC2 phosphorylation. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410416 Cd Length: 52 Bit Score: 39.74 E-value: 1.98e-04
|
||||||||||
| EFh | smart00054 | EF-hand, calcium binding motif; EF-hands are calcium-binding motifs that occur at least in ... |
161-184 | 2.57e-04 | ||||||
EF-hand, calcium binding motif; EF-hands are calcium-binding motifs that occur at least in pairs. Links between disease states and genes encoding EF-hands, particularly the S100 subclass, are emerging. Each motif consists of a 12 residue loop flanked on either side by a 12 residue alpha-helix. EF-hands undergo a conformational change unpon binding calcium ions. Pssm-ID: 197492 [Multi-domain] Cd Length: 29 Bit Score: 38.51 E-value: 2.57e-04
|
||||||||||
| C1 | smart00109 | Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol ... |
271-320 | 2.60e-04 | ||||||
Protein kinase C conserved region 1 (C1) domains (Cysteine-rich domains); Some bind phorbol esters and diacylglycerol. Some bind RasGTP. Zinc-binding domains. Pssm-ID: 197519 Cd Length: 50 Bit Score: 39.37 E-value: 2.60e-04
|
||||||||||
| C1_PDZD8 | cd20825 | protein kinase C conserved region 1 (C1 domain) found in PDZ domain-containing protein 8 ... |
204-252 | 2.71e-04 | ||||||
protein kinase C conserved region 1 (C1 domain) found in PDZ domain-containing protein 8 (PDZD8) and similar proteins; PDZD8, also called Sarcoma antigen NY-SAR-84/NY-SAR-104, is a molecular tethering protein that connects endoplasmic reticulum (ER) and mitochondrial membranes. PDZD8-dependent ER-mitochondria membrane tethering is essential for ER-mitochondria Ca2+ transfer. In neurons, it is involved in the regulation of dendritic Ca2+ dynamics by regulating mitochondrial Ca2+ uptake. PDZD8 also plays an indirect role in the regulation of cell morphology and cytoskeletal organization. It contains a PDZ domain and a C1 domain. This model describes the C1 domain, a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410375 Cd Length: 55 Bit Score: 39.18 E-value: 2.71e-04
|
||||||||||
| PRK13057 | PRK13057 | lipid kinase; |
423-467 | 3.34e-04 | ||||||
lipid kinase; Pssm-ID: 183857 [Multi-domain] Cd Length: 287 Bit Score: 43.37 E-value: 3.34e-04
|
||||||||||
| C1_TNS1_v | cd20888 | protein kinase C conserved region 1 (C1 domain) found in tensin-1 (TNS1) variant and similar ... |
207-254 | 5.38e-04 | ||||||
protein kinase C conserved region 1 (C1 domain) found in tensin-1 (TNS1) variant and similar proteins; Tensin-1 (TNS1) plays a role in fibrillar adhesion formation. It may be involved in cell migration, cartilage development and in linking signal transduction pathways to the cytoskeleton. This model corresponds to the C1 domain found in TNS1 variant. Typical TNS1 does not contain C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410438 Cd Length: 57 Bit Score: 38.70 E-value: 5.38e-04
|
||||||||||
| EF-hand_1 | pfam00036 | EF hand; The EF-hands can be divided into two classes: signalling proteins and buffering ... |
160-184 | 7.60e-04 | ||||||
EF hand; The EF-hands can be divided into two classes: signalling proteins and buffering/transport proteins. The first group is the largest and includes the most well-known members of the family such as calmodulin, troponin C and S100B. These proteins typically undergo a calcium-dependent conformational change which opens a target binding site. The latter group is represented by calbindin D9k and do not undergo calcium dependent conformational changes. Pssm-ID: 425435 [Multi-domain] Cd Length: 29 Bit Score: 37.38 E-value: 7.60e-04
|
||||||||||
| C1_Stac | cd20817 | protein kinase C conserved region 1 (C1 domain) found in the SH3 and cysteine-rich ... |
207-254 | 1.18e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the SH3 and cysteine-rich domain-containing protein (Stac) family; Stac proteins are putative adaptor proteins that are important for neuronal function. There are three mammalian members (Stac1, Stac2 and Stac3) of this family. Stac1 and Stac3 contain two SH3 domains while Stac2 contains a single SH3 domain at the C-terminus. Stac1 and Stac2 have been found to be expressed differently in mature dorsal root ganglia (DRG) neurons. Stac1 is mainly expressed in peptidergic neurons while Stac2 is found in a subset of nonpeptidergic and all trkB+ neurons. Stac proteins contain a cysteine-rich C1 domain and one or two SH3 domains at the C-terminus. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410367 Cd Length: 51 Bit Score: 37.31 E-value: 1.18e-03
|
||||||||||
| C1_MRCK | cd20809 | protein kinase C conserved region 1 (C1 domain) found in the Myotonic dystrophy kinase-related ... |
207-257 | 1.52e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the Myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) family; MRCK is thought to be a coincidence detector of signaling by the small GTPase Cdc42 and phosphoinositides. MRCK/Cdc42 signaling mediates myosin-dependent cell motility. MRCK has been shown to promote cytoskeletal reorganization, which affects many biological processes. Three isoforms of MRCK are known, named alpha, beta and gamma. MRCKgamma is expressed in heart and skeletal muscles, unlike MRCKalpha and MRCKbeta, which are expressed ubiquitously. MRCK consists of a serine/threonine kinase domain, a cysteine rich (C1) region, a PH domain and a p21 binding motif. This model corresponds to C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410359 Cd Length: 53 Bit Score: 37.25 E-value: 1.52e-03
|
||||||||||
| C1_Stac2 | cd20881 | protein kinase C conserved region 1 (C1 domain) found in SH3 and cysteine-rich ... |
207-254 | 1.77e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in SH3 and cysteine-rich domain-containing protein 2 (Stac2) and similar proteins; Stac2, also called 24b2/Stac2, or Src homology 3 and cysteine-rich domain-containing protein 2, plays a redundant role in promoting the expression of calcium channel CACNA1S at the cell membrane, and thereby contributes to increased channel activity. It slows down the inactivation rate of the calcium channel CACNA1C. Stac2 contains a cysteine-rich C1 domain and one SH3 domain at the C-terminus. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410431 Cd Length: 59 Bit Score: 37.12 E-value: 1.77e-03
|
||||||||||
| C1_TNS2 | cd20887 | protein kinase C conserved region 1 (C1 domain) found in tensin-2 and similar proteins; ... |
207-253 | 1.78e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in tensin-2 and similar proteins; Tensin-2 (TNS2), also called C1 domain-containing phosphatase and tensin (C1-TEN), or tensin-like C1 domain-containing phosphatase (TENC1), is an essential component for the maintenance of glomerular basement membrane (GBM) structures. It regulates cell motility and proliferation. It may have phosphatase activity. TNS2 reduces AKT1 phosphorylation, lowers AKT1 kinase activity, and interferes with AKT1 signaling. It contains an N-terminal region with a zinc finger (C1 domain), a protein tyrosine phosphatase (PTP)-like domain and a protein kinase 2 (C2) domain, and a C-terminal region with SH2 and pTyr binding (PTB) domains. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410437 Cd Length: 53 Bit Score: 37.07 E-value: 1.78e-03
|
||||||||||
| C1_DGKepsilon_typeIII_rpt1 | cd20801 | first protein kinase C conserved region 1 (C1 domain) found in type III diacylglycerol kinase, ... |
207-251 | 2.65e-03 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in type III diacylglycerol kinase, DAG kinase epsilon, and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase epsilon, also called diglyceride kinase epsilon (DGK-epsilon), is the only isoform classified as type III; it possesses a hydrophobic domain in addition to C1 and catalytic domains that are present in all DGKs, and shows selectivity for acyl chains. It is highly selective for arachidonate-containing species of DAG. It may terminate signals transmitted through arachidonoyl-DAG or may contribute to the synthesis of phospholipids with defined fatty acid composition. DAG kinase epsilon contains two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410351 Cd Length: 54 Bit Score: 36.53 E-value: 2.65e-03
|
||||||||||
| C1_cPKC_rpt1 | cd20833 | first protein kinase C conserved region 1 (C1 domain) found in the classical (or conventional) ... |
206-247 | 4.49e-03 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in the classical (or conventional) protein kinase C (cPKC) family; PKCs are classified into three groups (classical, atypical, and novel) depending on their mode of activation and the structural characteristics of their regulatory domains. cPKCs are potent kinases for histones, myelin basic protein, and protamine. They depend on calcium, DAG (1,2-diacylglycerol), and in most cases, phosphatidylserine (PS) for activation. There are four cPKC isoforms, named alpha, betaI, betaII, and gamma. PKC-alpha is expressed in many tissues and is associated with cell proliferation, apoptosis, and cell motility. It plays a role in the signaling of the growth factors PDGF, VEGF, EGF, and FGF. Abnormal levels of PKC-alpha have been detected in many transformed cell lines and several human tumors. In addition, PKC-alpha is required for HER2 dependent breast cancer invasion. The PKC beta isoforms (I and II), generated by alternative splicing of a single gene, are preferentially activated by hyperglycemia-induced DAG (1,2-diacylglycerol) in retinal tissues. This is implicated in diabetic microangiopathy such as ischemia, neovascularization, and abnormal vasodilator function. PKC-beta also plays an important role in VEGF signaling. In addition, glucose regulates proliferation in retinal endothelial cells via PKC-betaI. PKC-beta is also being explored as a therapeutic target in cancer. It contributes to tumor formation and is involved in the tumor host mechanisms of inflammation and angiogenesis. PKC-gamma is mainly expressed in neuronal tissues. It plays a role in protection from ischemia. Members of this family contain two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410383 Cd Length: 58 Bit Score: 35.85 E-value: 4.49e-03
|
||||||||||
| C1_ARHGEF-like | cd20832 | protein kinase C conserved region 1 (C1 domain) found in uncharacterized Rho guanine ... |
206-247 | 4.77e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in uncharacterized Rho guanine nucleotide exchange factor (ARHGEF)-like proteins; The family includes a group of uncharacterized proteins that show high sequence similarity to vertebrate Rho guanine nucleotide exchange factors ARHGEF11 and ARHGEF12, which may play a role in the regulation of RhoA GTPase by guanine nucleotide-binding alpha-12 (GNA12) and alpha-13 (GNA13). Unlike typical ARHGEF11 and ARHGEF12, members of this family contain a C1 domain. This model corresponds to the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410382 Cd Length: 53 Bit Score: 35.81 E-value: 4.77e-03
|
||||||||||
| C1_DGK_typeII_rpt2 | cd20852 | second protein kinase C conserved region 1 (C1 domain) found in type II diacylglycerol kinases; ... |
271-320 | 6.04e-03 | ||||||
second protein kinase C conserved region 1 (C1 domain) found in type II diacylglycerol kinases; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. Type II DAG kinases (DGKs) contain pleckstrin homology (PH) and sterile alpha motifs (SAM) domains, in addition to C1 and catalytic domains that are present in all DGKs. The SAM domain mediates oligomerization of type II DGKs. Three DGK isozymes (delta, eta and kappa) are classified as type II. DAG kinase delta, also called 130 kDa DAG kinase, or diglyceride kinase delta (DGK-delta), is a residential lipid kinase in the endoplasmic reticulum. It promotes lipogenesis and is involved in triglyceride biosynthesis. DAG kinase eta, also called diglyceride kinase eta (DGK-eta), plays a key role in promoting cell growth. The DAG kinase eta gene, DGKH, is a replicated risk gene of bipolar disorder (BPD). DAG kinase kappa is also called diglyceride kinase kappa (DGK-kappa) or 142 kDa DAG kinase. Members of this family contain two copies of the C1 domain. This model corresponds to the second one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410402 Cd Length: 54 Bit Score: 35.38 E-value: 6.04e-03
|
||||||||||
| C1_DGKtheta_typeV_rpt1 | cd20803 | first protein kinase C conserved region 1 (C1 domain) found in type V diacylglycerol kinase, ... |
207-249 | 6.05e-03 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in type V diacylglycerol kinase, DAG kinase theta, and similar proteins; Diacylglycerol (DAG) kinase (EC 2.7.1.107) is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid. DAG kinase theta, also called diglyceride kinase theta (DGK-theta), is the only isoform classified as type V; it contains a pleckstrin homology (PH)-like domain and an additional C1 domain, compared to other DGKs. It may regulate the activity of protein kinase C by controlling the balance between the two signaling lipids, diacylglycerol and phosphatidic acid. DAG kinase theta contains three copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410353 Cd Length: 56 Bit Score: 35.36 E-value: 6.05e-03
|
||||||||||
| PRK12361 | PRK12361 | hypothetical protein; Provisional |
424-466 | 6.78e-03 | ||||||
hypothetical protein; Provisional Pssm-ID: 183473 [Multi-domain] Cd Length: 547 Bit Score: 39.60 E-value: 6.78e-03
|
||||||||||
| C1_Raf | cd20811 | protein kinase C conserved region 1 (C1 domain) found in the Raf (Rapidly Accelerated ... |
206-255 | 6.83e-03 | ||||||
protein kinase C conserved region 1 (C1 domain) found in the Raf (Rapidly Accelerated Fibrosarcoma) kinase family; Raf kinases are serine/threonine kinases (STKs) that catalyze the transfer of the gamma-phosphoryl group from ATP to serine/threonine residues on protein substrates. They act as mitogen-activated protein kinase kinase kinases (MAP3Ks, MKKKs, MAPKKKs), which phosphorylate and activate MAPK kinases (MAPKKs or MKKs or MAP2Ks), which in turn phosphorylate and activate MAPKs during signaling cascades that are important in mediating cellular responses to extracellular signals. They function in the linear Ras-Raf-MEK-ERK pathway that regulates many cellular processes including cycle regulation, proliferation, differentiation, survival, and apoptosis. Aberrant expression or activation of components in this pathway are associated with tumor initiation, progression, and metastasis. Raf proteins contain a Ras binding domain, a zinc finger cysteine-rich domain (C1), and a catalytic kinase domain. Vertebrates have three Raf isoforms (A-, B-, and C-Raf) with different expression profiles, modes of regulation, and abilities to function in the ERK cascade, depending on cellular context and stimuli. They have essential and non-overlapping roles during embryo- and organogenesis. Knockout of each isoform results in a lethal phenotype or abnormality in most mouse strains. This model describes the C1 domain. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410361 Cd Length: 49 Bit Score: 35.35 E-value: 6.83e-03
|
||||||||||
| C1_cPKC_nPKC_rpt1 | cd20792 | first protein kinase C conserved region 1 (C1 domain) found in classical (or conventional) ... |
206-247 | 7.43e-03 | ||||||
first protein kinase C conserved region 1 (C1 domain) found in classical (or conventional) protein kinase C (cPKC), novel protein kinase C (nPKC), and similar proteins; PKCs are classified into three groups (classical, atypical, and novel) depending on their mode of activation and the structural characteristics of their regulatory domains. PKCs undergo three phosphorylations in order to take mature forms. In addition, cPKCs depend on calcium, DAG (1,2-diacylglycerol), and in most cases, phosphatidylserine (PS) for activation. nPKCs are calcium-independent, but require DAG and PS for activity, while atypical PKCs (aPKCs) only require PS. PKCs phosphorylate and modify the activities of a wide variety of cellular proteins including receptors, enzymes, cytoskeletal proteins, transcription factors, and other kinases. They play a central role in signal transduction pathways that regulate cell migration and polarity, proliferation, differentiation, and apoptosis. This family includes classical PKCs (cPKCs) and novel PKCs (nPKCs). There are four cPKC isoforms (named alpha, betaI, betaII, and gamma) and four nPKC isoforms (delta, epsilon, eta, and theta). Members of this family contain two copies of the C1 domain. This model corresponds to the first one. The C1 domain is a cysteine-rich zinc binding domain that does not bind DNA nor possess structural similarity to conventional zinc finger domains; it contains two separate Zn(2+)-binding sites. Pssm-ID: 410342 Cd Length: 53 Bit Score: 35.30 E-value: 7.43e-03
|
||||||||||
| EFh_PEF_ALG-2_like | cd16185 | EF-hand, calcium binding motif, found in homologs of mammalian apoptosis-linked gene 2 protein ... |
106-183 | 8.91e-03 | ||||||
EF-hand, calcium binding motif, found in homologs of mammalian apoptosis-linked gene 2 protein (ALG-2); The family includes some homologs of mammalian apoptosis-linked gene 2 protein (ALG-2) mainly found in lower eukaryotes, such as a parasitic protist Leishmarua major and a cellular slime mold Dictyostelium discoideum. These homologs contains five EF-hand motifs. Due to the presence of unfavorable residues at the Ca2+-coordinating positions, their non-canonical EF4 and EF5 hands may not bind Ca2+. Two Dictyostelium PEF proteins are the prototypes of this family. They may bind to cytoskeletal proteins and/or signal-transducing proteins localized to detergent-resistant membranes named lipid rafts, and occur as monomers or weak homo- or heterodimers like ALG-2. They can serve as a mediator for Ca2+ signaling-related Dictyostehum programmed cell death (PCD). Pssm-ID: 320060 [Multi-domain] Cd Length: 163 Bit Score: 37.58 E-value: 8.91e-03
|
||||||||||
| Blast search parameters | ||||
|