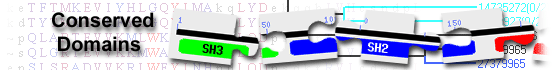granulocyte-macrophage colony-stimulating factor receptor subunit alpha isoform h [Homo sapiens]
fibronectin type III domain-containing protein( domain architecture ID 229438)
fibronectin type III (FN3) domain-containing protein may be involved in specific interactions with other molecules through its FN3 domain; similar to Homo sapiens interleukin-5 receptor subunit alpha and interferon gamma receptor 2
List of domain hits

| Name | Accession | Description | Interval | E-value | |||
| FN3 super family | cl21522 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein ... |
1-82 | 6.31e-22 | |||
Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. The actual alignment was detected with superfamily member pfam09240: Pssm-ID: 473895 Cd Length: 96 Bit Score: 87.08 E-value: 6.31e-22
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| IL6Ra-bind | pfam09240 | Interleukin-6 receptor alpha chain, binding; Members of this family adopt a structure ... |
1-82 | 6.31e-22 | |||
Interleukin-6 receptor alpha chain, binding; Members of this family adopt a structure consisting of an immunoglobulin-like beta-sandwich, with seven strands in two beta-sheets, in a Greek-key topology. They are required for binding to the cytokine Interleukin-6. Pssm-ID: 401252 Cd Length: 96 Bit Score: 87.08 E-value: 6.31e-22
|
|||||||
| Name | Accession | Description | Interval | E-value | |||
| IL6Ra-bind | pfam09240 | Interleukin-6 receptor alpha chain, binding; Members of this family adopt a structure ... |
1-82 | 6.31e-22 | |||
Interleukin-6 receptor alpha chain, binding; Members of this family adopt a structure consisting of an immunoglobulin-like beta-sandwich, with seven strands in two beta-sheets, in a Greek-key topology. They are required for binding to the cytokine Interleukin-6. Pssm-ID: 401252 Cd Length: 96 Bit Score: 87.08 E-value: 6.31e-22
|
|||||||
| Blast search parameters | ||||
|